Carbohydrate Ligands for COVID-19 Spike Proteins
- PMID: 35215921
- PMCID: PMC8880561
- DOI: 10.3390/v14020330
Carbohydrate Ligands for COVID-19 Spike Proteins
Abstract
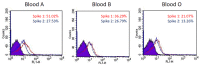
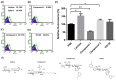
An outbreak of SARS-CoV-2 coronavirus (COVID-19) first detected in Wuhan, China, has created a public health emergency all over the world. The pandemic has caused more than 340 million confirmed cases and 5.57 million deaths as of 23 January 2022. Although carbohydrates have been found to play a role in coronavirus binding and infection, the role of cell surface glycans in SARS-CoV-2 infection and pathogenesis is still not understood. Herein, we report that the SARS-CoV-2 spike protein S1 subunit binds specifically to blood group A and B antigens, and that the spike protein S2 subunit has a binding preference for Lea antigens. Further examination of the binding preference for different types of red blood cells (RBCs) indicated that the spike protein S1 subunit preferentially binds with blood group A RBCs, whereas the spike protein S2 subunit prefers to interact with blood group Lea RBCs. Angiotensin converting enzyme 2 (ACE2), a known target of SARS-CoV-2 spike proteins, was identified to be a blood group A antigen-containing glycoprotein. Additionally, 6-sulfo N-acetyllactosamine was found to inhibit the binding of the spike protein S1 subunit with blood group A RBCs and reduce the interaction between the spike protein S1 subunit and ACE2.
Keywords: COVID-19; blood group; carbohydrate ligand; spike protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

