Dynamics of HIV-1 Gag Processing as Revealed by Fluorescence Lifetime Imaging Microscopy and Single Virus Tracking
- PMID: 35215933
- PMCID: PMC8874525
- DOI: 10.3390/v14020340
Dynamics of HIV-1 Gag Processing as Revealed by Fluorescence Lifetime Imaging Microscopy and Single Virus Tracking
Abstract
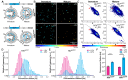
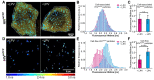
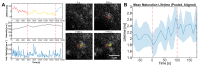
The viral polyprotein Gag plays a central role for HIV-1 assembly, release and maturation. Proteolytic processing of Gag by the viral protease is essential for the structural rearrangements that mark the transition from immature to mature, infectious viruses. The timing and kinetics of Gag processing are not fully understood. Here, fluorescence lifetime imaging microscopy and single virus tracking are used to follow Gag processing in nascent HIV-1 particles in situ. Using a Gag polyprotein labelled internally with eCFP, we show that proteolytic release of the fluorophore from Gag is accompanied by an increase in its fluorescence lifetime. By tracking nascent virus particles in situ and analyzing the intensity and fluorescence lifetime of individual traces, we detect proteolytic cleavage of eCFP from Gag in a subset (6.5%) of viral particles. This suggests that for the majority of VLPs, Gag processing occurs with a delay after particle assembly.
Keywords: HIV; fluorescence lifetime; gag processing; maturation; single virus tracking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Pettit S.C., Moody M.D., Wehbie R.S., Kaplan A.H., Nantermet P.V., Klein C.A., Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

