Molecular Characterization and Pathogenicity of a Novel Soybean-Infecting Monopartite Geminivirus in China
- PMID: 35215936
- PMCID: PMC8877103
- DOI: 10.3390/v14020341
Molecular Characterization and Pathogenicity of a Novel Soybean-Infecting Monopartite Geminivirus in China
Abstract
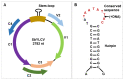
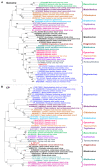
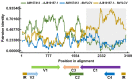
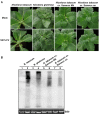
Soybean is a major legume crop that plays an important role in food production, industrial production, and animal husbandry. Here, we characterize a novel soybean-infecting monopartite geminivirus identified in China. Analysis of the contigs de novo assembled from sequenced small interfering RNAs, followed by PCR, cloning, and sequencing, the complete viral genome was determined to be 2782 nucleotides. The genome contains the conserved nonanucleotide sequence, TAATATTAC and other sequence features typical of the family Geminiviridae, and encodes two and four open reading frames in the virion-sense and the complementary-sense strands, respectively. Genome-wide pairwise identity analysis revealed that the novel virus shares less than 65.6% identity with previously characterized geminiviruses. Phylogenetic and recombination analysis indicated that this virus was placed in a unique taxon within the family Geminiviridae and potentially arose from recombination. An infectious clone of this virus was further constructed and its infectivity was tested in different species of plants. Successful infection and characteristic symptoms were observed in Glycine max, Nicotiana benthamiana, N. tabacum, N. glutinosa, and N. tabacum cv. Samsun plants. Taken together, this virus represents a member of an unclassified genus of the family Geminiviridae, for which the name soybean yellow leaf curl virus is proposed.
Keywords: geminivirus; recombination; soybean; ssDNA virus.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, orin the decision to publish the results.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

