Structure-Based Discovery of N-Sulfonylpiperidine-3-Carboxamides as Novel Capsid Assembly Modulators for Potent Inhibition of HBV Replication
- PMID: 35215939
- PMCID: PMC8876525
- DOI: 10.3390/v14020348
Structure-Based Discovery of N-Sulfonylpiperidine-3-Carboxamides as Novel Capsid Assembly Modulators for Potent Inhibition of HBV Replication
Abstract
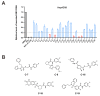
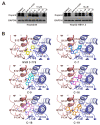
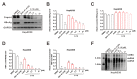
As a key element during HBV replication, a nucleocapsid is considered a promising target for the treatment of chronic hepatitis B. The present study aimed to identify small molecules as novel capsid assembly modulators with antiviral activity. Structure-based virtual screening of an integrated compound library led to the identification of several types of HBV inhibitors. Among these inhibitors, N-sulfonylpiperidine-3-carboxamides (SPCs) potently reduced the amount of secreted HBV DNA. Through structure-activity relationship studies, we identified an SPC derivative, namely, C-39, which exhibited the highest antiviral activity without causing cytotoxicity. Mechanism studies showed that C-39 dose-dependently inhibited the formation of HBV capsid, synthesis of cccDNA, e antigen (HBeAg), viral pregenomic RNA (pgRNA), and HBV DNA levels, thereby restraining HBV replication. In summary, SPCs represent a new class of capsid assembly modulators. Further optimization of SPCs is expected to obtain new antiviral drugs against HBV infection.
Keywords: antiviral activity; assembly modulator; capsid; core protein; hepatitis B virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Novel Potent Capsid Assembly Modulators Regulate Multiple Steps of the Hepatitis B Virus Life Cycle.Antimicrob Agents Chemother. 2018 Sep 24;62(10):e00835-18. doi: 10.1128/AAC.00835-18. Print 2018 Oct. Antimicrob Agents Chemother. 2018. PMID: 30012770 Free PMC article.
-
Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids.J Virol. 2013 Jun;87(12):6931-42. doi: 10.1128/JVI.00582-13. Epub 2013 Apr 10. J Virol. 2013. PMID: 23576513 Free PMC article.
-
Hepatitis B Virus Capsid Assembly Modulators, but Not Nucleoside Analogs, Inhibit the Production of Extracellular Pregenomic RNA and Spliced RNA Variants.Antimicrob Agents Chemother. 2017 Jul 25;61(8):e00680-17. doi: 10.1128/AAC.00680-17. Print 2017 Aug. Antimicrob Agents Chemother. 2017. PMID: 28559265 Free PMC article.
-
Treatment of Chronic Hepatitis B Virus Infection Using Small Molecule Modulators of Nucleocapsid Assembly: Recent Advances and Perspectives.ACS Infect Dis. 2019 May 10;5(5):713-724. doi: 10.1021/acsinfecdis.8b00337. Epub 2019 Mar 29. ACS Infect Dis. 2019. PMID: 30896149 Review.
-
Small Molecule Inhibitors of Hepatitis B Virus Nucleocapsid Assembly: A New Approach to Treat Chronic HBV Infection.Curr Med Chem. 2018;25(7):802-813. doi: 10.2174/0929867324666170704121800. Curr Med Chem. 2018. PMID: 28675991 Review.
References
-
- Razavi-Shearer D., Gamkrelidze I., Nguyen M.H., Chen D.-S., Van Damme P., Abbas Z., Abdulla M., Abou Rached A., Adda D., Aho I., et al. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: A modelling study. Lancet Gastroenterol. Hepatol. 2018;3:383–403. doi: 10.1016/S2468-1253(18)30056-6. - DOI - PubMed
-
- World Health Organization Hepatitis B Fact Sheet. [(accessed on 27 July 2020)]. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

