Zika Virus Infection of Sertoli Cells Alters Protein Expression Involved in Activated Immune and Antiviral Response Pathways, Carbohydrate Metabolism and Cardiovascular Disease
- PMID: 35215967
- PMCID: PMC8878972
- DOI: 10.3390/v14020377
Zika Virus Infection of Sertoli Cells Alters Protein Expression Involved in Activated Immune and Antiviral Response Pathways, Carbohydrate Metabolism and Cardiovascular Disease
Abstract
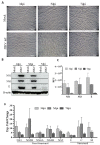
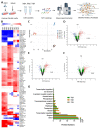
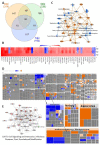
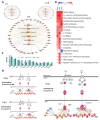
Zika virus (ZIKV), a re-emerging virus, causes congenital brain abnormalities and Guillain-Barré syndrome. It is mainly transmitted by Aedes mosquitoes, but infections are also linked to sexual transmissions. Infectious ZIKV has been isolated, and viral RNA has been detected in semen over a year after the onset of initial symptoms, but the mode of long-term persistence is not yet understood. ZIKV can proliferate in human Sertoli cells (HSerC) for several weeks in vitro, suggesting that it might be a reservoir for persistent ZIKV infection. This study determined proteomic changes in HSerC during ZIKV infections by TMT-mass spectrometry analysis. Levels of 4416 unique Sertoli cell proteins were significantly altered at 3, 5, and 7 days after ZIKV infection. The significantly altered proteins include enzymes, transcription regulators, transporters, kinases, peptidases, transmembrane receptors, cytokines, ion channels, and growth factors. Many of these proteins are involved in pathways associated with antiviral response, antigen presentation, and immune cell activation. Several immune response pathway proteins were significantly activated during infection, e.g., interferon signaling, T cell receptor signaling, IL-8 signaling, and Th1 signaling. The altered protein levels were linked to predicted activation of immune response in HSerC, which was predicted to suppress ZIKV infection. ZIKV infection also affected the levels of critical regulators of gluconeogenesis and glycolysis pathways such as phosphoglycerate mutase, phosphoglycerate kinase, and enolase. Interestingly, many significantly altered proteins were associated with cardiac hypertrophy, which may induce heart failure in infected patients. In summary, our research contributes to a better understanding of ZIKV replication dynamics and infection in Sertoli cells.
Keywords: Sertoli cells; Zika virus; carbohydrate metabolism; cardiovascular disease; glycolysis; immune response; persistence in semen; sexual transmission.
Conflict of interest statement
The authors declare they have no conflicts of interest with this study.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

