Unlike Chloroquine, Mefloquine Inhibits SARS-CoV-2 Infection in Physiologically Relevant Cells
- PMID: 35215969
- PMCID: PMC8874959
- DOI: 10.3390/v14020374
Unlike Chloroquine, Mefloquine Inhibits SARS-CoV-2 Infection in Physiologically Relevant Cells
Abstract
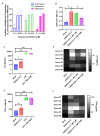
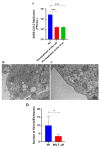
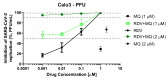
Despite the development of specific therapies against severe acute respiratory coronavirus 2 (SARS-CoV-2), the continuous investigation of the mechanism of action of clinically approved drugs could provide new information on the druggable steps of virus-host interaction. For example, chloroquine (CQ)/hydroxychloroquine (HCQ) lacks in vitro activity against SARS-CoV-2 in TMPRSS2-expressing cells, such as human pneumocyte cell line Calu-3, and likewise, failed to show clinical benefit in the Solidarity and Recovery clinical trials. Another antimalarial drug, mefloquine, which is not a 4-aminoquinoline like CQ/HCQ, has emerged as a potential anti-SARS-CoV-2 antiviral in vitro and has also been previously repurposed for respiratory diseases. Here, we investigated the anti-SARS-CoV-2 mechanism of action of mefloquine in cells relevant for the physiopathology of COVID-19, such as Calu-3 cells (that recapitulate type II pneumocytes) and monocytes. Molecular pathways modulated by mefloquine were assessed by differential expression analysis, and confirmed by biological assays. A PBPK model was developed to assess mefloquine's optimal doses for achieving therapeutic concentrations. Mefloquine inhibited SARS-CoV-2 replication in Calu-3, with an EC50 of 1.2 µM and EC90 of 5.3 µM. It reduced SARS-CoV-2 RNA levels in monocytes and prevented virus-induced enhancement of IL-6 and TNF-α. Mefloquine reduced SARS-CoV-2 entry and synergized with Remdesivir. Mefloquine's pharmacological parameters are consistent with its plasma exposure in humans and its tissue-to-plasma predicted coefficient points suggesting that mefloquine may accumulate in the lungs. Altogether, our data indicate that mefloquine's chemical structure could represent an orally available host-acting agent to inhibit virus entry.
Keywords: COVID-19; SARS-CoV-2; antimalarial drug; antiviral; mefloquine.
Conflict of interest statement
The authors declare no conflict of interest and the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- World Health Organization . WHO R&D Blueprint: Informal Consultation on Prioritization of Candidate Therapeutic Agents for Use in Novel Coronavirus 2019 Infection. WHO; Geneva, Switzerland: 2020.
-
- PubMed Identifying SARS-CoV-2-Related Coronaviruses in Malayan Pangolins. [(accessed on 20 October 2020)]; Available online: https://pubmed.ncbi.nlm.nih.gov/32218527/ - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

