An Update of Orthopoxvirus Molecular Evolution
- PMID: 35215981
- PMCID: PMC8875945
- DOI: 10.3390/v14020388
An Update of Orthopoxvirus Molecular Evolution
Abstract
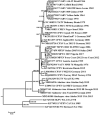
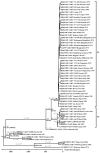
Although variola virus (VARV) has been eradicated through widespread vaccination, other orthopoxviruses pathogenic for humans circulate in nature. Recently, new orthopoxviruses, including some able to infect humans, have been found and their complete genomes have been sequenced. Questions about the orthopoxvirus mutation rate and the emergence of new threats to humankind as a result of the evolution of circulating orthopoxviruses remain open. Based on contemporary data on ancient VARV DNA and DNA of new orthopoxvirus species, an analysis of the molecular evolution of orthopoxviruses was carried out and the timescale of their emergence was estimated. It was calculated that the orthopoxviruses of the Old and New Worlds separated approximately 40,000 years ago; the recently discovered Akhmeta virus and Alaskapox virus separated from other orthopoxviruses approximately 10,000-20,000 years ago; the rest of modern orthopoxvirus species originated from 1700 to 6000 years ago, with the exception of VARV, which emerged in approximately 300 AD. Later, there was a separation of genetic variants of some orthopoxvirus species, so the monkeypox virus West African subtype originated approximately 600 years ago, and the VARV minor alastrim subtype emerged approximately 300 years ago.
Keywords: evolution; origin; orthopoxvirus; variola virus.
Conflict of interest statement
All co-authors have seen and agree with the contents of the manuscript and the order of authors, and there is no financial interest to report. All co-authors declare that they have no conflict of interest.
Figures


References
-
- Fenner F., Henderson D.A., Arita I., Jezek Z., Ladnyi I.D. Smallpox and Its Eradication. WHO; Geneva, Switzerland: 1988.
-
- Mühlemann B., Vinner L., Margaryan A., Wilhelmson H., de la Fuente Castro C., Allentoft M.E., de Barros Damgaard P., Hansen A.J., Holtsmark Nielsen S., Strand L., et al. Diverse variola virus (smallpox) strains were widespread in northern Europe in the Viking Age. Science. 2020;369:eaaw8977. doi: 10.1126/science.aaw8977. - DOI - PubMed
-
- Shchelkunov S.N., Marennikova S.S., Moyer R.W. Orthopoxviruses Pathogenic for Humans. Springer; Berlin/Heildeberg Germany: 2005.
-
- Chen N., Bellone C.J., Schriewer J., Owens G., Fredrickson T., Parker S., Buller R.M.L. Poxvirus interleukin-4 expression overcomes inherent resistance and vaccine-induced immunity: Pathogenesis, prophylaxis, and antiviral therapy. Virology. 2011;409:328–337. doi: 10.1016/j.virol.2010.10.021. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

