Sensitivity of SARS-CoV-2 Life Cycle to IFN Effects and ACE2 Binding Unveiled with a Stochastic Model
- PMID: 35215996
- PMCID: PMC8875829
- DOI: 10.3390/v14020403
Sensitivity of SARS-CoV-2 Life Cycle to IFN Effects and ACE2 Binding Unveiled with a Stochastic Model
Abstract
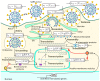
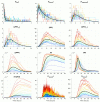
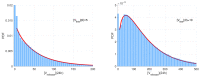
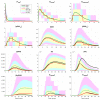
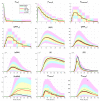
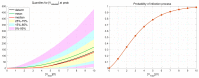
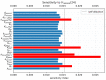
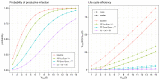
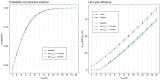
Mathematical modelling of infection processes in cells is of fundamental interest. It helps to understand the SARS-CoV-2 dynamics in detail and can be useful to define the vulnerability steps targeted by antiviral treatments. We previously developed a deterministic mathematical model of the SARS-CoV-2 life cycle in a single cell. Despite answering many questions, it certainly cannot accurately account for the stochastic nature of an infection process caused by natural fluctuation in reaction kinetics and the small abundance of participating components in a single cell. In the present work, this deterministic model is transformed into a stochastic one based on a Markov Chain Monte Carlo (MCMC) method. This model is employed to compute statistical characteristics of the SARS-CoV-2 life cycle including the probability for a non-degenerate infection process. Varying parameters of the model enables us to unveil the inhibitory effects of IFN and the effects of the ACE2 binding affinity. The simulation results show that the type I IFN response has a very strong effect on inhibition of the total viral progeny whereas the effect of a 10-fold variation of the binding rate to ACE2 turns out to be negligible for the probability of infection and viral production.
Keywords: Markov Chain Monte Carlo method; SARS-Cov-2; mathematical model; sensitivity analysis; stochastic processes; the ACE2 receptor; type I interferon (IFN); virus dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ostaszewski M., Niarakis A., Mazein A., Kuperstein I., Phair R., Orta-Resendiz A., Singh V., Aghamiri S.S., Acencio M.L., Glaab E., et al. COVID-19 Disease Map Community, a computational knowledge repository of virus-host interaction mechanisms. Mol. Syst. Biol. 2021;17:e10387. doi: 10.15252/msb.202110387. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

