Collection of Monoclonal Antibodies Targeting SARS-CoV-2 Proteins
- PMID: 35216036
- PMCID: PMC8875891
- DOI: 10.3390/v14020443
Collection of Monoclonal Antibodies Targeting SARS-CoV-2 Proteins
Abstract
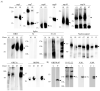
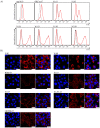
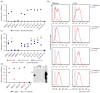
In early 2020, the COVID-19 pandemic sparked a global crisis that continues to pose a serious threat to human health and the economy. Further advancement in research is necessary and requires the availability of quality molecular tools, including monoclonal antibodies. Here, we present the development and characterization of a collection of over 40 new monoclonal antibodies directed against different SARS-CoV-2 proteins. Recombinant SARS-CoV-2 proteins were expressed, purified, and used as immunogens. Upon development of specific hybridomas, the obtained monoclonal antibody (mAb) clones were tested for binding to recombinant proteins and infected cells. We generated mAbs against structural proteins, the Spike and Nucleocapsid protein, several non-structural proteins (nsp1, nsp7, nsp8, nsp9, nsp10, nsp16) and accessory factors (ORF3a, ORF9b) applicable in flow cytometry, immunofluorescence, or Western blot. Our collection of mAbs provides a set of novel, highly specific tools that will allow a comprehensive analysis of the viral proteome, which will allow further understanding of SARS-CoV-2 pathogenesis and the design of therapeutic strategies.
Keywords: COVID-19; SARS-CoV-2; monoclonal antibodies; variants of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

