Root Membrane Ubiquitinome under Short-Term Osmotic Stress
- PMID: 35216074
- PMCID: PMC8879470
- DOI: 10.3390/ijms23041956
Root Membrane Ubiquitinome under Short-Term Osmotic Stress
Abstract
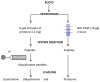
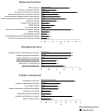
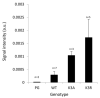
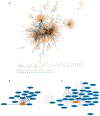
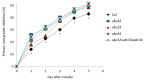
Osmotic stress can be detrimental to plants, whose survival relies heavily on proteomic plasticity. Protein ubiquitination is a central post-translational modification in osmotic-mediated stress. In this study, we used the K-Ɛ-GG antibody enrichment method integrated with high-resolution mass spectrometry to compile a list of 719 ubiquitinated lysine (K-Ub) residues from 450 Arabidopsis root membrane proteins (58% of which are transmembrane proteins), thereby adding to the database of ubiquitinated substrates in plants. Although no ubiquitin (Ub) motifs could be identified, the presence of acidic residues close to K-Ub was revealed. Our ubiquitinome analysis pointed to a broad role of ubiquitination in the internalization and sorting of cargo proteins. Moreover, the simultaneous proteome and ubiquitinome quantification showed that ubiquitination is mostly not involved in membrane protein degradation in response to short osmotic treatment but that it is putatively involved in protein internalization, as described for the aquaporin PIP2;1. Our in silico analysis of ubiquitinated proteins shows that two E2 Ub-conjugating enzymes, UBC32 and UBC34, putatively target membrane proteins under osmotic stress. Finally, we revealed a positive role for UBC32 and UBC34 in primary root growth under osmotic stress.
Keywords: aquaporin; mass spectrometry; osmotic stress; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

