The dual role of p62 in ferroptosis of glioblastoma according to p53 status
- PMID: 35216629
- PMCID: PMC8881833
- DOI: 10.1186/s13578-022-00764-z
The dual role of p62 in ferroptosis of glioblastoma according to p53 status
Abstract
Background: Ferroptosis plays a key role in human cancer, but its function and mechanism in glioma is not clear. P62/SQSTM1 was reported to inhibit ferroptosis via the activation of NRF2 signaling pathway. In this study we reveal a dual role of p62 in ferroptosis of glioblastoma (GBM) according to p53 status.
Method: Lipid peroxidation analysis, transmission electron microscopy (TEM), GSH assay were performed to determine the level of ferroptosis. Western blot and qPCR were obtained to detect the expression of ferroptosis markers. Construction of mutant plasmids, immunoprecipitation, luciferase assay and rescue-experiments were performed to explore the regulatory mechanism.
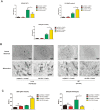
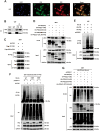
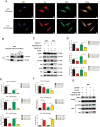
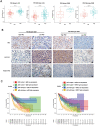
Results: P62 overexpression facilitates ferroptosis and inhibits SLC7A11 expression in p53 mutant GBM, while attenuates ferroptosis and promotes SLC7A11 expression in p53 wild-type GBM. P62 associates with p53 and inhibits its ubiquitination. The p53-NRF2 association and p53-mediated suppression of NRF2 antioxidant activity are diversely regulated by p62 according to p53 status. P53 mutation status is required for the dual regulation of p62 on ferroptosis. In wild-type p53 GBM, the classical p62-mediated NRF2 activation pathway plays a major regulatory role of ferroptosis, leading to increased SLC7A11 expression, resulting in a anti-ferroptosis role. In mutant p53 GBM, stronger interaction of mutant-p53/NRF2 by p62 enhance the inhibitory effect of mutant p53 on NRF2 signaling, which reversing the classical p62-mediated NRF2 activation pathway, together with increased p53's transcriptional suppression on SLC7A11 by p62, leading to a decrease of SLC7A11, resulting in a pro-ferroptosis role.
Conclusion: Together, this study shows novel molecular mechanisms of ferroptosis regulated by p62; the mutation status of p53 is an important factor that determines the therapeutic response to p62-mediated ferroptosis-targeted therapies in GBM.
Keywords: Ferroptosis; Glioblastoma; NRF2; p53; p62.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Rho family GTPase 1 (RND1), a novel regulator of p53, enhances ferroptosis in glioblastoma.Cell Biosci. 2022 May 3;12(1):53. doi: 10.1186/s13578-022-00791-w. Cell Biosci. 2022. PMID: 35505371 Free PMC article.
-
SLC7A11 regulated by NRF2 modulates esophageal squamous cell carcinoma radiosensitivity by inhibiting ferroptosis.J Transl Med. 2021 Aug 26;19(1):367. doi: 10.1186/s12967-021-03042-7. J Transl Med. 2021. PMID: 34446045 Free PMC article.
-
CircLRFN5 inhibits the progression of glioblastoma via PRRX2/GCH1 mediated ferroptosis.J Exp Clin Cancer Res. 2022 Oct 20;41(1):307. doi: 10.1186/s13046-022-02518-8. J Exp Clin Cancer Res. 2022. PMID: 36266731 Free PMC article.
-
Role of ferroptosis on tumor progression and immunotherapy.Cell Death Discov. 2022 Oct 26;8(1):427. doi: 10.1038/s41420-022-01218-8. Cell Death Discov. 2022. PMID: 36289191 Free PMC article. Review.
-
Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy.Protein Cell. 2021 Aug;12(8):599-620. doi: 10.1007/s13238-020-00789-5. Epub 2020 Oct 1. Protein Cell. 2021. PMID: 33000412 Free PMC article. Review.
Cited by
-
Propranolol and Capecitabine Synergy on Inducing Ferroptosis in Human Colorectal Cancer Cells: Potential Implications in Cancer Therapy.Cancers (Basel). 2025 Apr 27;17(9):1470. doi: 10.3390/cancers17091470. Cancers (Basel). 2025. PMID: 40361395 Free PMC article.
-
TP63 as a modulator of ferroptosis in TP53 mutations glioblastoma.Cell Death Dis. 2025 Aug 13;16(1):614. doi: 10.1038/s41419-025-07938-w. Cell Death Dis. 2025. PMID: 40796737 Free PMC article.
-
Pyroptosis, ferroptosis, and autophagy cross-talk in glioblastoma opens up new avenues for glioblastoma treatment.Cell Commun Signal. 2023 May 19;21(1):115. doi: 10.1186/s12964-023-01108-1. Cell Commun Signal. 2023. PMID: 37208730 Free PMC article. Review.
-
Post-Translational Modifications of p53 in Ferroptosis: Novel Pharmacological Targets for Cancer Therapy.Front Pharmacol. 2022 May 24;13:908772. doi: 10.3389/fphar.2022.908772. eCollection 2022. Front Pharmacol. 2022. PMID: 35685623 Free PMC article. Review.
-
An oncogene regulating chromatin favors response to immunotherapy: Oncogene CHAF1A and immunotherapy outcomes.Oncoimmunology. 2024 Jan 9;13(1):2303195. doi: 10.1080/2162402X.2024.2303195. eCollection 2024. Oncoimmunology. 2024. PMID: 38235318 Free PMC article.
References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. - PubMed
-
- Stockwell BR, Friedmann AJ, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis: a regulated cell death nexus linking metabolism. Redox Biol Dis Cell. 2017;171:273–285. - PMC - PubMed
-
- Su Y, Zhao B, Zhou L, Zhang Z, Shen Y, Lv H, AlQudsy L, Shang P. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett. 2020;483:127–136. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

