Multiple early factors anticipate post-acute COVID-19 sequelae
- PMID: 35216672
- PMCID: PMC8786632
- DOI: 10.1016/j.cell.2022.01.014
Multiple early factors anticipate post-acute COVID-19 sequelae
Abstract
Post-acute sequelae of COVID-19 (PASC) represent an emerging global crisis. However, quantifiable risk factors for PASC and their biological associations are poorly resolved. We executed a deep multi-omic, longitudinal investigation of 309 COVID-19 patients from initial diagnosis to convalescence (2-3 months later), integrated with clinical data and patient-reported symptoms. We resolved four PASC-anticipating risk factors at the time of initial COVID-19 diagnosis: type 2 diabetes, SARS-CoV-2 RNAemia, Epstein-Barr virus viremia, and specific auto-antibodies. In patients with gastrointestinal PASC, SARS-CoV-2-specific and CMV-specific CD8+ T cells exhibited unique dynamics during recovery from COVID-19. Analysis of symptom-associated immunological signatures revealed coordinated immunity polarization into four endotypes, exhibiting divergent acute severity and PASC. We find that immunological associations between PASC factors diminish over time, leading to distinct convalescent immune states. Detectability of most PASC factors at COVID-19 diagnosis emphasizes the importance of early disease measurements for understanding emergent chronic conditions and suggests PASC treatment strategies.
Keywords: COVID-19; PASC; RNAemia; antibodies; auto-antibodies; computational biology; immune system; immunology; long COVID; metabolomics; multi-omics; proteomics; single cell; single-cell CITE-seq; single-cell RNA-seq; single-cell TCR-seq; single-cell secretome; symptoms; transcriptome; viremia.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.R.H. and A.R. are founders and board members of PACT Pharma. J.R.H. is a board member of Isoplexis, and A.R. is the scientific advisor to Isoplexis. M.M.D. is a member of the Scientific Advisory Board of PACT Pharma. J.A.B. is a member of the Scientific Advisory Boards of Arcus, Solid, and VIR. J.A.B. is a member of the Board of Directors of Gilead and Provention. J.A.B. is the CEO of Sonoma Biotherapeutics. L.L.L. is on the scientific advisory boards of Alector, Atreca, Dragonfly, DrenBio, Nkarta, Obsidian Therapeutics, and SBI Biotech. R.G. has received consulting income from Juno Therapeutics, Takeda, Infotech Soft, Celgene, and Merck, has received research support from Janssen Pharmaceuticals and Juno Therapeutics, and declares ownership in CellSpace Biosciences. P.D.G. is on the Scientific Advisory Board of Celsius, Earli, Elpiscience, Immunoscape, Rapt, and Nextech, was a scientific founder of Juno Therapeutics, and receives research support from Lonza. J.D.G. declared contracted research with Gilead, Lilly, and Regeneron. J.A.H. received consulting fees or honoraria from Gilead Sciences, Amplyx, Allovir, Allogene therapeutics, CRISPR therapeutics, CSL Behring, OptumHealth, Octapharma, and Takeda and research funding from Takeda, Allovir, Karius, and Gilead Sciences. Q.D., D.H.M., R.T., R.M.G., M.K., and T.M.S. have employment and equity ownership with Adaptive Biotechnologies. The remaining authors declare no competing interests.
Figures



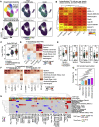
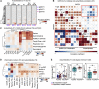



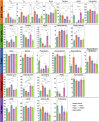

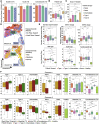

References
-
- Annunziato F., Romagnani C., Romagnani S. The 3 major types of innate and adaptive cell-mediated effector immunity. J. Allergy Clin. Immunol. 2015;135:626–635. - PubMed
-
- Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., Michailidis E., Hoffmann H.H., Eto S., Garcia-Prat M., et al. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci. Immunol. 2021;6 eabl4340. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007290/AI/NIAID NIH HHS/United States
- T32 AI007334/AI/NIAID NIH HHS/United States
- R21 AI138258/AI/NIAID NIH HHS/United States
- R01 DA040395/DA/NIDA NIH HHS/United States
- UG3 TR002884/TR/NCATS NIH HHS/United States
- R01 AI068129/AI/NIAID NIH HHS/United States
- P01 AI153559/AI/NIAID NIH HHS/United States
- R01 AI155685/AI/NIAID NIH HHS/United States
- T32 AI007044/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U19 AI128914/AI/NIAID NIH HHS/United States
- R01 AI163131/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

