SARS-CoV-2 spike antigen quantification by targeted mass spectrometry of a virus-based vaccine
- PMID: 35217103
- PMCID: PMC8863330
- DOI: 10.1016/j.jviromet.2022.114498
SARS-CoV-2 spike antigen quantification by targeted mass spectrometry of a virus-based vaccine
Abstract
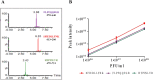
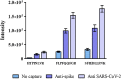
The spike glycoprotein mediates virus binding to the host cells and is a key target for vaccines development. One SARS-CoV-2 vaccine is based on vesicular stomatitis virus (VSV), in which the native surface glycoprotein has been replaced by the SARS-CoV-2 spike protein (VSV-ΔG-spike). The titer of the virus is quantified by the plaque forming unit (PFU) assay, but there is no method for spike protein quantitation as an antigen in a VSV-based vaccine. Here, we describe a mass spectrometric (MS) spike protein quantification method, applied to VSV-ΔG-spike based vaccine. Proof of concept of this method, combining two different sample preparations, is shown for complex matrix samples, produced during the vaccine manufacturing processes. Total spike levels were correlated with results from activity assays, and ranged between 0.3-0.5 μg of spike protein per 107 PFU virus-based vaccine. This method is simple, linear over a wide range, allows quantification of antigen within a sample and can be easily implemented for any vaccine or therapeutic sample.
Keywords: Mass spectrometry; Quantification; SARS-CoV-2; Spike; Vaccine.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Barlev-Gross M., Weiss S., Ben-Shmuel A., Sittner A., Eden K., Mazuz N., Glinert I., Bar-David E., Puni R., Amit S., Kriger O., Schuster O., Alcalay R., Makdasi E., Epstein E., Noy-Porat T., Rosenfeld R., Achdout H., Mazor O., Israely T., Levy H., Mechaly A. Spike vs nucleocapsid SARS-CoV-2 antigen detection: application in nasopharyngeal swab specimens. Anal. Bioanal. Chem. 2021;413:3501–3510. doi: 10.1007/s00216-021-03298-4. - DOI - PMC - PubMed
-
- Gouveia D., Miotello G., Gallais F., Gaillard J.-C., Debroas S., Bellanger L., Lavigne J.-P., Sotto A., Grenga L., Pible O., Armengaud J. Proteotyping SARS-CoV-2 virus from nasopharyngeal swabs: a proof-of-Concept focused on a 3 min mass spectrometry window. J. Proteome Res. 2020;19:4407–4416. doi: 10.1021/acs.jproteome.0c00535. - DOI - PubMed
-
- Lerer E., Oren Z., Kafri Y., Adar Y., Cherry L., Lupu E., Monash A., Levy R., Dor E., Epstein E., Levin L., Girshengorn M., Hazan O., Simon I., Tal A., Tzadoka H., Makovitzki A. Highly efficient purification of recombinant VSV-ΔG-spike vaccine against SARS-CoV-2 by flow-through chromatography. BioTech. 2021;10 doi: 10.3390/biotech10040022. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

