Differential requirements for different subfamilies of the mammalian SWI/SNF chromatin remodeling enzymes in myoblast cell cycle progression and expression of the Pax7 regulator
- PMID: 35217218
- PMCID: PMC8948540
- DOI: 10.1016/j.bbagrm.2022.194801
Differential requirements for different subfamilies of the mammalian SWI/SNF chromatin remodeling enzymes in myoblast cell cycle progression and expression of the Pax7 regulator
Abstract
The mammalian SWItch/Sucrose Non-Fermentable (mSWI/SNF) families of ATP-dependent chromatin remodeling enzymes are established co-regulators of gene expression. mSWI/SNF complexes can be assembled into three major subfamilies: BAF (BRG1 or BRM-Associated Factor), PBAF (Polybromo containing BAF), or ncBAF (non-canonical BAF) that are distinguished by the presence of mutually exclusive subunits. The mechanisms by which each subfamily contributes to the establishment or function of specific cell lineages are poorly understood. Here, we determined the contributions of the BAF, ncBAF, and PBAF complexes to myoblast proliferation via knock down (KD) of distinguishing subunits from each complex. KD of subunits unique to the BAF or the ncBAF complexes reduced myoblast proliferation rate, while KD of PBAF-specific subunits did not affect proliferation. RNA-seq from proliferating KD myoblasts targeting Baf250A (BAF complex), Brd9 (ncBAF complex), or Baf180 (PBAF complex) showed mis-regulation of a limited number of genes. KD of Baf250A specifically reduced the expression of Pax7, which is required for myoblast proliferation, concomitant with decreased binding of Baf250A to and impaired chromatin remodeling at the Pax7 gene promoter. Although Brd9 also bound to the Pax7 promoter, suggesting occupancy by the ncBAF complex, no changes were detected in Pax7 gene expression, Pax7 protein expression or chromatin remodeling at the Pax7 promoter upon Brd9 KD. The data indicate that the BAF subfamily of the mSWI/SNF enzymes is specifically required for myoblast proliferation via regulation of Pax7 expression.
Keywords: Baf250A; Myoblasts; Pax7; Proliferation; SWI/SNF.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
ETHICS DECLARATIONS
The authors declare no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
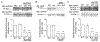
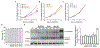



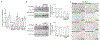


Similar articles
-
Differential Contributions of mSWI/SNF Chromatin Remodeler Sub-Families to Myoblast Differentiation.Int J Mol Sci. 2023 Jul 9;24(14):11256. doi: 10.3390/ijms241411256. Int J Mol Sci. 2023. PMID: 37511016 Free PMC article.
-
Identification of assembly mode of non-canonical BAF (ncBAF) chromatin remodeling complex core module.Biochem Biophys Res Commun. 2025 Jan;745:151238. doi: 10.1016/j.bbrc.2024.151238. Epub 2024 Dec 25. Biochem Biophys Res Commun. 2025. PMID: 39732119
-
Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes.Cell. 2018 Nov 15;175(5):1272-1288.e20. doi: 10.1016/j.cell.2018.09.032. Epub 2018 Oct 18. Cell. 2018. PMID: 30343899 Free PMC article.
-
Preclinical evidence in the assembly of mammalian SWI/SNF complexes: Epigenetic insights and clinical perspectives in human lung disease therapy.Mol Ther. 2024 Aug 7;32(8):2470-2488. doi: 10.1016/j.ymthe.2024.06.026. Epub 2024 Jun 22. Mol Ther. 2024. PMID: 38910326 Free PMC article. Review.
-
The role of the polybromo-associated BAF complex in development.Biochem Cell Biol. 2025 Jan 1;103:1-8. doi: 10.1139/bcb-2024-0224. Epub 2024 Nov 14. Biochem Cell Biol. 2025. PMID: 39541575 Free PMC article. Review.
Cited by
-
Cysteine Rich Intestinal Protein 2 is a copper-responsive regulator of skeletal muscle differentiation and metal homeostasis.PLoS Genet. 2024 Dec 5;20(12):e1011495. doi: 10.1371/journal.pgen.1011495. eCollection 2024 Dec. PLoS Genet. 2024. PMID: 39637238 Free PMC article.
-
Functional effect of indole-3 carbinol in the viability and invasive properties of cultured cancer cells.Biochem Biophys Rep. 2023 Jun 1;35:101492. doi: 10.1016/j.bbrep.2023.101492. eCollection 2023 Sep. Biochem Biophys Rep. 2023. PMID: 37304131 Free PMC article.
-
Muscle-specific pyruvate kinase isoforms, PKM1 and PKM2, regulate mammalian SWI/SNF proteins and histone 3 phosphorylation during myoblast differentiation.FASEB J. 2024 Jun 15;38(11):e23702. doi: 10.1096/fj.202400784R. FASEB J. 2024. PMID: 38837439 Free PMC article.
-
Muscle-Specific Pyruvate Kinase Isoforms, Pkm1 and Pkm2, Regulate Mammalian SWI/SNF Proteins and Histone 3 Phosphorylation During Myoblast Differentiation.bioRxiv [Preprint]. 2024 Apr 11:2024.04.10.588959. doi: 10.1101/2024.04.10.588959. bioRxiv. 2024. Update in: FASEB J. 2024 Jun 15;38(11):e23702. doi: 10.1096/fj.202400784R. PMID: 38645038 Free PMC article. Updated. Preprint.
-
Cysteine Rich Intestinal Protein 2 is a copper-responsive regulator of skeletal muscle differentiation.bioRxiv [Preprint]. 2024 May 5:2024.05.03.592485. doi: 10.1101/2024.05.03.592485. bioRxiv. 2024. Update in: PLoS Genet. 2024 Dec 5;20(12):e1011495. doi: 10.1371/journal.pgen.1011495. PMID: 38746126 Free PMC article. Updated. Preprint.
References
-
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR, Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex, Nature, 370 (1994) 477–481. - PubMed
-
- Imbalzano AN, Kwon H, Green MR, Kingston RE, Facilitated binding of TATA-binding protein to nucleosomal DNA, Nature, 370 (1994) 481–485. - PubMed
-
- Cote J, Quinn J, Workman JL, Peterson CL, Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex, Science, 265 (1994) 53–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

