Evolutionarily conserved inhibitory uORFs sensitize Hox mRNA translation to start codon selection stringency
- PMID: 35217614
- PMCID: PMC8892498
- DOI: 10.1073/pnas.2117226119
Evolutionarily conserved inhibitory uORFs sensitize Hox mRNA translation to start codon selection stringency
Abstract
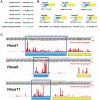
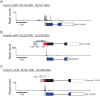
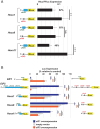
Translation start site selection in eukaryotes is influenced by context nucleotides flanking the AUG codon and by levels of the eukaryotic translation initiation factors eIF1 and eIF5. In a search of mammalian genes, we identified five homeobox (Hox) gene paralogs initiated by AUG codons in conserved suboptimal context as well as 13 Hox genes that contain evolutionarily conserved upstream open reading frames (uORFs) that initiate at AUG codons in poor sequence context. An analysis of published cap analysis of gene expression sequencing (CAGE-seq) data and generated CAGE-seq data for messenger RNAs (mRNAs) from mouse somites revealed that the 5' leaders of Hox mRNAs of interest contain conserved uORFs, are generally much shorter than reported, and lack previously proposed internal ribosome entry site elements. We show that the conserved uORFs inhibit Hox reporter expression and that altering the stringency of start codon selection by overexpressing eIF1 or eIF5 modulates the expression of Hox reporters. We also show that modifying ribosome homeostasis by depleting a large ribosomal subunit protein or treating cells with sublethal concentrations of puromycin leads to lower stringency of start codon selection. Thus, altering global translation can confer gene-specific effects through altered start codon selection stringency.
Keywords: Hox genes; eIF1; eIF5; start codon selection stringency; uORF.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5.Nucleic Acids Res. 2012 Apr;40(7):2898-906. doi: 10.1093/nar/gkr1192. Epub 2011 Dec 7. Nucleic Acids Res. 2012. PMID: 22156057 Free PMC article.
-
Translational regulation by uORFs and start codon selection stringency.Genes Dev. 2023 Jun 1;37(11-12):474-489. doi: 10.1101/gad.350752.123. Epub 2023 Jul 11. Genes Dev. 2023. PMID: 37433636 Free PMC article. Review.
-
eIF1 discriminates against suboptimal initiation sites to prevent excessive uORF translation genome-wide.RNA. 2020 Apr;26(4):419-438. doi: 10.1261/rna.073536.119. Epub 2020 Jan 8. RNA. 2020. PMID: 31915290 Free PMC article.
-
Translational autoregulation of BZW1 and BZW2 expression by modulating the stringency of start codon selection.PLoS One. 2018 Feb 22;13(2):e0192648. doi: 10.1371/journal.pone.0192648. eCollection 2018. PLoS One. 2018. PMID: 29470543 Free PMC article.
-
A helicase links upstream ORFs and RNA structure.Curr Genet. 2019 Apr;65(2):453-456. doi: 10.1007/s00294-018-0911-z. Epub 2018 Nov 27. Curr Genet. 2019. PMID: 30483885 Free PMC article. Review.
Cited by
-
RpL38 modulates germ cell differentiation by controlling Bam expression in Drosophila testis.Sci China Life Sci. 2024 Nov;67(11):2411-2425. doi: 10.1007/s11427-024-2646-3. Epub 2024 Aug 21. Sci China Life Sci. 2024. PMID: 39187660
-
Chaperone-directed ribosome repair after oxidative damage.Mol Cell. 2023 May 4;83(9):1527-1537.e5. doi: 10.1016/j.molcel.2023.03.030. Epub 2023 Apr 21. Mol Cell. 2023. PMID: 37086725 Free PMC article.
-
The molecular basis of translation initiation and its regulation in eukaryotes.Nat Rev Mol Cell Biol. 2024 Mar;25(3):168-186. doi: 10.1038/s41580-023-00624-9. Epub 2023 Dec 5. Nat Rev Mol Cell Biol. 2024. PMID: 38052923 Review.
-
Ribosome biogenesis and function in development and disease.Development. 2023 Mar 1;150(5):dev201187. doi: 10.1242/dev.201187. Epub 2023 Mar 7. Development. 2023. PMID: 36897354 Free PMC article. Review.
-
N-terminal alanine-rich (NTAR) sequences drive precise start codon selection resulting in elevated translation of multiple proteins including ERK1/2.Nucleic Acids Res. 2023 Aug 25;51(15):7714-7735. doi: 10.1093/nar/gkad528. Nucleic Acids Res. 2023. PMID: 37414542 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

