Natural variation in Glume Coverage 1 causes naked grains in sorghum
- PMID: 35217660
- PMCID: PMC8881591
- DOI: 10.1038/s41467-022-28680-3
Natural variation in Glume Coverage 1 causes naked grains in sorghum
Abstract
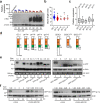
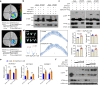
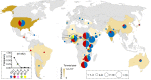
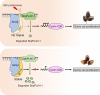
One of the most critical steps in cereal threshing is the ease with which seeds are detached from sticky glumes. Naked grains with low glume coverage have dramatically increased threshing efficiency and seed quality. Here, we demonstrate that GC1 (Glume Coverage 1), encoding an atypical G protein γ subunit, negatively regulates sorghum glume coverage. Naturally truncated variations of GC1 C-terminus accumulate at higher protein levels and affect the stability of a patatin-related phospholipase SbpPLAII-1. A strong positive selection signature around the GC1 genic region is found in the naked sorghum cultivars. Our findings reveal a crucial event during sorghum domestication through a subtle regulation of glume development by GC1 C-terminus variation, and establish a strategy for future breeding of naked grains.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Fine mapping of qGW1, a major QTL for grain weight in sorghum.Theor Appl Genet. 2015 Sep;128(9):1813-25. doi: 10.1007/s00122-015-2549-2. Epub 2015 Jun 13. Theor Appl Genet. 2015. PMID: 26071275
-
Novel Grain Weight Loci Revealed in a Cross between Cultivated and Wild Sorghum.Plant Genome. 2018 Jul;11(2). doi: 10.3835/plantgenome2017.10.0089. Plant Genome. 2018. PMID: 30025022
-
Genetic architecture of kernel composition in global sorghum germplasm.BMC Genomics. 2017 Jan 5;18(1):15. doi: 10.1186/s12864-016-3403-x. BMC Genomics. 2017. PMID: 28056770 Free PMC article.
-
Genetic Architecture of Grain Yield-Related Traits in Sorghum and Maize.Int J Mol Sci. 2022 Feb 22;23(5):2405. doi: 10.3390/ijms23052405. Int J Mol Sci. 2022. PMID: 35269548 Free PMC article. Review.
-
Evolution of cereal floral architecture and threshability.Trends Plant Sci. 2023 Dec;28(12):1438-1450. doi: 10.1016/j.tplants.2023.08.003. Epub 2023 Sep 4. Trends Plant Sci. 2023. PMID: 37673701 Review.
Cited by
-
A high-quality chromosome-level genome assembly of Pelteobagrus vachelli provides insights into its environmental adaptation and population history.Front Genet. 2022 Nov 14;13:1050192. doi: 10.3389/fgene.2022.1050192. eCollection 2022. Front Genet. 2022. PMID: 36452160 Free PMC article.
-
R2R3 MYB transcription factor SbMYBHv33 negatively regulates sorghum biomass accumulation and salt tolerance.Theor Appl Genet. 2023 Jan;136(1):5. doi: 10.1007/s00122-023-04292-3. Epub 2023 Jan 19. Theor Appl Genet. 2023. PMID: 36656365
-
Knockout of a gene encoding a Gγ protein boosts alkaline tolerance in cereal crops.aBIOTECH. 2023 Jul 2;4(2):180-183. doi: 10.1007/s42994-023-00106-8. eCollection 2023 Jun. aBIOTECH. 2023. PMID: 37581019 Free PMC article.
-
Genetic control of morphological traits useful for improving sorghum.Breed Sci. 2023 Mar;73(1):57-69. doi: 10.1270/jsbbs.22069. Epub 2023 Jan 17. Breed Sci. 2023. PMID: 37168813 Free PMC article.
-
Chromosomal inversion at the DG1 promoter drives double-grain spikelets and enhances grain yield in sorghum.Nat Plants. 2025 Mar;11(3):453-467. doi: 10.1038/s41477-025-01937-7. Epub 2025 Mar 11. Nat Plants. 2025. PMID: 40069576
References
-
- Adeyanju A, Perumal R, Tesso T. Genetic analysis of threshability in grain sorghum [Sorghum bicolor (L.) Moench] Plant Breed. 2015;134:148–155.
-
- Smith, W. C. & Frederiksen, R. A. Sorghum: Origin, History, Technology and Production (Wiley, 2000).
-
- Ayyangar GNR, Ayyar MAS, Narayanan TR, Nambiar AKK. The grain sorghums of the Durra group. Proc. Natl Acad. Sci. India Sect. B Biol. Sci. 1942;15:133–147.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

