A method comparison study of the high throughput automated HISCL® SARS-CoV-2 antigen assay using nasopharyngeal swab samples from symptomatic and asymptomatic subjects against conventional RT-PCR
- PMID: 35218042
- PMCID: PMC9088525
- DOI: 10.1002/jmv.27679
A method comparison study of the high throughput automated HISCL® SARS-CoV-2 antigen assay using nasopharyngeal swab samples from symptomatic and asymptomatic subjects against conventional RT-PCR
Abstract
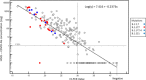
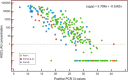
Our study aim was to evaluate the performance of the automated Sysmex HISCL® severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen assay against reverse-transcription polymerase chain reaction (RT-PCR). We tested 277 remnant frozen nasopharyngeal swab samples, stored in universal transport medium (UTM), yielding a sensitivity of 94.9% against historical RT-PCR results with cycle threshold (Ct ) < 30, and a sensitivity of 76.7% for Ct < 35, and specificity of 100% (all Ct values) confirming compatibility of UTM-diluted samples with the assay system. Thereafter, we prospectively collected 141 nasopharyngeal swab samples in UTM from healthcare workers and 1369 paired swabs (400 UTM; 969 dry) from individuals at a public health testing center, with the first swab (UTM) reserved for RT-PCR, yielding a positivity rate of 4.6%. HISCL assay performance using UTM swabs was superior to dry swabs, with a sensitivity of 100% (95% confidence interval [CI] 71.5%-100%) at Ct < 30 versus 92.3% (95%CI 81.5%-97.9%), and a specificity of 99.3% (95% CI 98.1-99.89) against 83.3% (95%CI 80.7%-85.6%). We conclude that this antigen assay is suitable for high throughput facilities where the primary indication for testing is to rule out infection with low RT-PCR Ct values (proxy for high viral loads) to curb viral spread.
Keywords: HISCL automated antigen assay; RT-PCR; SARS-CoV-2; method comparison; rapid testing; variants of concern.
© 2022 The Authors. Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
Joachim Linssen and Marion Münster are full‐time employees of Sysmex Europe GMBH who funded the study. The other authors have no conflicts of interest to declare.
Figures

References
-
- Ghandour HHS, Meara J, McClain C. BMJ Global Health. BMJ Publishing Group Limited; 2020. https://blogs.bmj.com/bmjgh/2020/06/04/lessons-from-the-covid-19-pandemi...
-
- Corman VBT, Brünink S, Drosten C, Landt O, Koopmans M, Zambon M. Diagnostic Detection of 2019‐nCoV by Real‐Time RT‐PCR. World Health Organization; 2020.
-
- WHO . Emergency Use Listing for In vitro diagnostics (IVDs) Detecting SARS‐CoV‐2. World Health Organization; 2021. https://extranet.who.int/pqweb/news/who-emergency-use-listing-vitro-diag...
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

