The spike-ACE2 binding assay: An in vitro platform for evaluating vaccination efficacy and for screening SARS-CoV-2 inhibitors and neutralizing antibodies
- PMID: 35218866
- PMCID: PMC8863957
- DOI: 10.1016/j.jim.2022.113244
The spike-ACE2 binding assay: An in vitro platform for evaluating vaccination efficacy and for screening SARS-CoV-2 inhibitors and neutralizing antibodies
Abstract
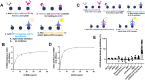
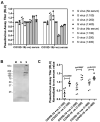
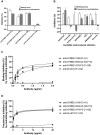
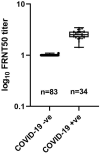
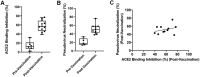
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 has become a worldwide pandemic, and there is a pressing need for the rapid development of novel therapeutic strategies. SARS-CoV-2 viral entry is mediated by interaction between the receptor binding domain (RBD) of the SARS-CoV-2 Spike protein and host cellular receptor, human angiotensin converting enzyme 2 (ACE2). The lack of a high throughput screening (HTS) platform for candidate drug screening means that no targeted COVID-19 treatments have been developed to date. To overcome this limitation, we developed a novel, rapid, simple, and HTS binding assay platform to screen potential inhibitors of the RBD-ACE2 complex. Three "neutralizing" mouse monoclonal antibodies capable of blocking the RBD-ACE2 interaction were identified using our binding assay and pseudovirus neutralization assay followed by further validation with the Focus Reduction Neutralization Test (FRNT), which analyzes the neutralization capacity of samples in the presence of live SARS-CoV-2. Furthermore, the consistency of our binding assay and FRNT results (R2 = 0.68) was demonstrated by patients' serum, of which were COVID-19 positive (n = 34) and COVID-19 negative (n = 76). Several small molecules selected for their potential to inhibit the Spike-ACE2 complex in silico were also confirmed with the binding assay. In addition, we have evaluated vaccine efficacy using binding assay platform and validated through pseudovirus neutralization assay. The correlation between binding assay & psuedovirus assay of the post vaccinated serum showed well correlated (R2 = 0.09) Moreover, our binding assay platform successfully validated different Spike RBD mutants. These results indicate that our binding assay can be used as a platform for in vitro screening of small molecules and monoclonal antibodies, and high-throughput assessment of antibody levels after vaccination. When conducting drug screening, computer virtual screening lacks actual basis, construction of pseudoviruses is relatively complicated, and even FRNT requires a P3 laboratory. There are few methods to determine the competitiveness of the target drug and SRBD or ACE2. Our binding assay can fill this gap and accelerate the process and efficiency of COVID-19 drug screening.
Keywords: ACE2; Binding assay; COVID-19; FRNT; Inhibitor screening; Neutralization antibody; RBD; SARS-CoV-2; Spike-mutant; Vaccine.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Shuhong Luo, Tuhin Das, Shuangzhe Zhang, Hao Tang, Xinyi Yao, Tarina Cho, Jingqiao Lu, Kino Maravillas, Valerie Jones, and Ruo-Pan Huang are employees of RayBiotech and have a financial stake in RayBiotech.
Figures

Similar articles
-
Competitive SARS-CoV-2 Serology Reveals Most Antibodies Targeting the Spike Receptor-Binding Domain Compete for ACE2 Binding.mSphere. 2020 Sep 16;5(5):e00802-20. doi: 10.1128/mSphere.00802-20. mSphere. 2020. PMID: 32938700 Free PMC article.
-
Correlation of SARS-CoV-2 Viral Neutralizing Antibody Titers with Anti-Spike Antibodies and ACE-2 Inhibition among Vaccinated Individuals.Microbiol Spectr. 2022 Oct 26;10(5):e0131522. doi: 10.1128/spectrum.01315-22. Epub 2022 Sep 19. Microbiol Spectr. 2022. PMID: 36121252 Free PMC article.
-
Characterization of a neutralizing antibody that recognizes a loop region adjacent to the receptor-binding interface of the SARS-CoV-2 spike receptor-binding domain.Microbiol Spectr. 2024 Apr 2;12(4):e0365523. doi: 10.1128/spectrum.03655-23. Epub 2024 Feb 28. Microbiol Spectr. 2024. PMID: 38415660 Free PMC article.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
-
Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies.Biochem Biophys Res Commun. 2021 Jan 29;538:192-203. doi: 10.1016/j.bbrc.2020.10.012. Epub 2020 Oct 10. Biochem Biophys Res Commun. 2021. PMID: 33069360 Free PMC article. Review.
Cited by
-
Immunological Studies to Understand Hybrid/Recombinant Variants of SARS-CoV-2.Vaccines (Basel). 2022 Dec 25;11(1):45. doi: 10.3390/vaccines11010045. Vaccines (Basel). 2022. PMID: 36679891 Free PMC article. Review.
-
N-glycosylation of the SARS-CoV-2 spike protein at Asn331 and Asn343 is involved in spike-ACE2 binding, virus entry, and regulation of IL-6.Microbiol Immunol. 2024 May;68(5):165-178. doi: 10.1111/1348-0421.13121. Epub 2024 Mar 6. Microbiol Immunol. 2024. PMID: 38444370 Free PMC article.
-
Decoy peptides effectively inhibit the binding of SARS-CoV-2 to ACE2 on oral epithelial cells.Heliyon. 2023 Nov 20;9(12):e22614. doi: 10.1016/j.heliyon.2023.e22614. eCollection 2023 Dec. Heliyon. 2023. PMID: 38107325 Free PMC article.
-
Overstated conclusions of a non-inferiority trial testing immunogenicity and safety of homologous and heterologous booster - authors' reply.Lancet Reg Health Southeast Asia. 2023 Apr 21;12:100204. doi: 10.1016/j.lansea.2023.100204. eCollection 2023 May. Lancet Reg Health Southeast Asia. 2023. PMID: 37384053 Free PMC article. No abstract available.
-
Establishment of a Yeast Two-Hybrid-Based High-Throughput Screening Model for Selection of SARS-CoV-2 Spike-ACE2 Interaction Inhibitors.Int J Mol Sci. 2025 Jan 15;26(2):678. doi: 10.3390/ijms26020678. Int J Mol Sci. 2025. PMID: 39859397 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

