IBD-associated G protein-coupled receptor 65 variant compromises signalling and impairs key functions involved in inflammation
- PMID: 35218908
- PMCID: PMC9536022
- DOI: 10.1016/j.cellsig.2022.110294
IBD-associated G protein-coupled receptor 65 variant compromises signalling and impairs key functions involved in inflammation
Abstract
Background and aims: Inflammatory bowel diseases (IBD) result in chronic inflammation of the gastrointestinal tract. Genetic studies have shown that the GPR65 gene, as well as its missense coding variant, GPR65*Ile231Leu, is associated with IBD. We aimed to define the signalling and biological pathways downstream of GPR65 activation and evaluate the impact of GPR65*231Leu on these.
Methods: We used HEK 293 cells stably expressing GPR65 and deficient for either Gαs, Gαq/11 or Gα12/13, to define GPR65 signalling pathways, IBD patient biopsies and a panel of human tissues, primary immune cells and cell lines to determine biologic context, and genetic modulation of human THP-1-derived macrophages to examine the impact of GPR65 in bacterial phagocytosis and NLRP3 inflammasome activation.
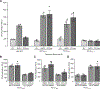
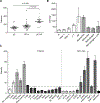
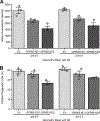
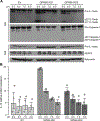
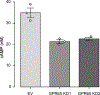
Results: We confirmed that GPR65 signals via the Gαs pathway, leading to cAMP accumulation. GPR65 can also signal via the Gα12/13 pathway leading to formation of stress fibers, actin remodeling and RhoA activation; all impaired by the IBD-associated GPR65*231Leu allele. Gene expression profiling revealed greater expression of GPR65 in biopsies from inflamed compared to non-inflamed tissues from IBD patients or control individuals, potentially explained by infiltration of inflammatory immune cells. Decreased GPR65 expression in THP-1-derived macrophages leads to impaired bacterial phagocytosis, increased NLRP3 inflammasome activation and IL-1β secretion in response to an inflammatory stimulus.
Conclusions: We demonstrate that GPR65 exerts its effects through Gαs- and Gα12/13-mediated pathways, that the IBD-associated GPR65*231Leu allele has compromised interactions with Gα12/13 and that KD of GPR65 leads to impaired bacterial phagocytosis and increased inflammatory signalling via the NLRP3 inflammasome. This work identifies a target for development of small molecule therapies.
Keywords: Actin remodeling; Bacterial phagocytosis; G proteins; GPR65; NLRP3 inflammasome.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest related to the contents of this article.
Figures
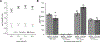

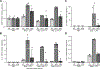
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

