Dynamic control of chromatin-associated m6A methylation regulates nascent RNA synthesis
- PMID: 35219383
- PMCID: PMC8969783
- DOI: 10.1016/j.molcel.2022.02.006
Dynamic control of chromatin-associated m6A methylation regulates nascent RNA synthesis
Abstract
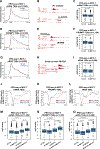
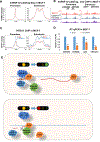
N6-methyladenosine (m6A) methylation is co-transcriptionally deposited on mRNA, but a possible role of m6A on transcription remains poorly understood. Here, we demonstrate that the METTL3/METTL14/WTAP m6A methyltransferase complex (MTC) is localized to many promoters and enhancers and deposits the m6A modification on nascent transcripts, including pre-mRNAs, promoter upstream transcripts (PROMPTs), and enhancer RNAs. PRO-seq analyses demonstrate that nascent RNAs originating from both promoters and enhancers are significantly decreased in the METTL3-depleted cells. Furthermore, genes targeted by the Integrator complex for premature termination are depleted of METTL3, suggesting a potential antagonistic relationship between METTL3 and Integrator. Consistently, we found the Integrator complex component INTS11 elevated at promoters and enhancers upon loss of MTC or nuclear m6A binders. Taken together, our findings suggest that MTC-mediated m6A modification protects nascent RNAs from Integrator-mediated termination and promotes productive transcription, thus unraveling an unexpected layer of gene regulation imposed by RNA m6A modification.
Keywords: ALKBH5; INTS11; METTL3; chromatin; enhancer; hnRNP G; m(6)A; nascent RNA; promoter.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Y.S. is a co-founder of and holds equity in K36 Therapeutics. Y.S. is a consultant for Active Motif, a member of the Scientific Advisory Board of the College of Life Sciences, West Lake University, and a member of the MD Anderson External Advisory Board. K.A. received research funding from Novartis not related to this work, is a consultant for Syros Pharmaceuticals, is on the SAB of CAMP4 Therapeutics, and is a member of the Advisory Board of Molecular Cell. All other authors declare no competing interests.
Figures


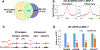

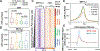
References
-
- Akhtar J, Renaud Y, Albrecht S, Ghavi-Helm Y, Roignant JY, Silies M, and Junion G (2021). m6A RNA methylation regulates promoter- proximal pausing of RNA polymerase II. MOL CELL 81, 3356–3367. - PubMed
-
- Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, and Nureki O, et al. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. SCIENCE 363. - PubMed
-
- Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, Dossin F, Fouassier C, Lameiras S, and Bourc’His D (2021). m(6)A RNA methylation regulates the fate of endogenous retroviruses. NATURE 591, 312–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

