Short-chain fatty acid acetate triggers antiviral response mediated by RIG-I in cells from infants with respiratory syncytial virus bronchiolitis
- PMID: 35220042
- PMCID: PMC8871125
- DOI: 10.1016/j.ebiom.2022.103891
Short-chain fatty acid acetate triggers antiviral response mediated by RIG-I in cells from infants with respiratory syncytial virus bronchiolitis
Abstract
Background: Gut microbiota-derived short-chain fatty-acid (SFCA) acetate protects mice against RSV A2 strain infection by increasing interferon-β production and expression of interferon-stimulated genes (ISGs). However, the role of SFCA in RSV infection using strains isolated from patients is unknown.
Methods: We first used RSV clinical strains isolated from infants hospitalized with RSV bronchiolitis to investigate the effects of in vitro SCFA-acetate treatment of human pulmonary epithelial cells. We next examined whether SCFA-acetate treatment is beneficial in a mouse model of RSV infection using clinical isolates. We sought to investigate the relationship of gut microbiota and fecal acetate with disease severity among infants hospitalized with RSV bronchiolitis, and whether treating their respiratory epithelial cells with SCFA-acetate ex-vivo impacts viral load and ISG expression. We further treated epithelial cells from SARS-CoV-2 infected patients with SCFA-acetate.
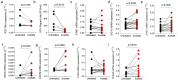
Findings: In vitro pre-treatment of A549 cells with SCFA-acetate reduced RSV infection with clinical isolates and increased the expression of RIG-I and ISG15. Animals treated with SCFA-acetate intranasally recovered significantly faster, with reduction in the RSV clinical isolates viral load, and increased lung expression of IFNB1 and the RIG-I. Experiments in RIG-I knockout A549 cells demonstrated that the protection relies on RIG-I presence. Gut microbial profile was associated with bronchiolitis severity and with acetate in stool. Increased SCFA-acetate levels were associated with increasing oxygen saturation at admission, and shorter duration of fever. Ex-vivo treatment of patients' respiratory cells with SCFA-acetate reduced RSV load and increased expression of ISGs OAS1 and ISG15, and virus recognition receptors MAVS and RIG-I, but not IFNB1. These SCFA-acetate effects were not found on cells from SARS-CoV-2 infected patients.
Interpretation: SCFA-acetate reduces the severity of RSV infection and RSV viral load through modulation of RIG-I expression.
Funding: FAPERGS (FAPERGS/MS/CNPq/SESRS no. 03/2017 - PPSUS 17/2551-0001380-8 and COVID-19 20/2551-0000258-6); CNPq 312504/2017-9; CAPES) - Finance Code 001.
Keywords: Acetate; Clinical isolates; RIG-I; Respiratory syncytial virus.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Foley D.A., Phuong L.K., Englund J.A. Respiratory syncytial virus immunisation overview. J Paediatr Child Health. 2020;56(12):1865–1867. - PubMed
-
- Mazur N.I., Higgins D., Nunes M.C., et al. The respiratory syncytial virus vaccine landscape: lessons from the graveyard and promising candidates. Lancet Infect Dis. 2018;18(10):e295–e311. - PubMed
-
- Ruckwardt T.J., Morabito K.M., Graham B.S. Immunological lessons from respiratory syncytial virus vaccine development. Immunity. 2019;51(3):429–442. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

