A novel role for nucleolin in splice site selection
- PMID: 35220879
- PMCID: PMC8890436
- DOI: 10.1080/15476286.2021.2020455
A novel role for nucleolin in splice site selection
Abstract
Latent 5' splice sites, not normally used, are highly abundant in human introns, but are activated under stress and in cancer, generating thousands of nonsense mRNAs. A previously proposed mechanism to suppress latent splicing was shown to be independent of NMD, with a pivotal role for initiator-tRNA independent of protein translation. To further elucidate this mechanism, we searched for nuclear proteins directly bound to initiator-tRNA. Starting with UV-crosslinking, we identified nucleolin (NCL) interacting directly and specifically with initiator-tRNA in the nucleus, but not in the cytoplasm. Next, we show the association of ini-tRNA and NCL with pre-mRNA. We further show that recovery of suppression of latent splicing by initiator-tRNA complementation is NCL dependent. Finally, upon nucleolin knockdown we show activation of latent splicing in hundreds of coding transcripts having important cellular functions. We thus propose nucleolin, a component of the endogenous spliceosome, through its direct binding to initiator-tRNA and its effect on latent splicing, as the first protein of a nuclear quality control mechanism regulating splice site selection to protect cells from latent splicing that can generate defective mRNAs.
Keywords: 5ʹ splice site selection; RNA sequencing; alternative splicing; bioinformatics analysis; endogenous spliceosome; latent splice sites; latent splicing; mass spectrometry; splicing regulation; suppression of splicing.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




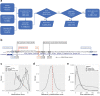


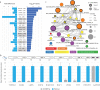

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
