A defective lysophosphatidic acid-autophagy axis increases miscarriage risk by restricting decidual macrophage residence
- PMID: 35220880
- PMCID: PMC9542369
- DOI: 10.1080/15548627.2022.2039000
A defective lysophosphatidic acid-autophagy axis increases miscarriage risk by restricting decidual macrophage residence
Abstract
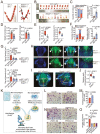
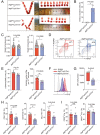
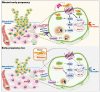
Massive infiltrated and enriched decidual macrophages (dMφ) have been widely regarded as important regulators of maternal-fetal immune tolerance and trophoblast invasion, contributing to normal pregnancy. However, the characteristics of metabolic profile and the underlying mechanism of dMφ residence remain largely unknown. Here, we observe that dMφ display an active glycerophospholipid metabolism. The activation of ENPP2-lysophosphatidic acid (LPA) facilitates the adhesion and retention, and M2 differentiation of dMφ during normal pregnancy. Mechanistically, this process is mediated through activation of the LPA receptors (LPAR1 and PPARG/PPARγ)-DDIT4-macroautophagy/autophagy axis, and further upregulation of multiple adhesion factors (e.g., cadherins and selectins) in a CLDN7 (claudin 7)-dependent manner. Additionally, poor trophoblast invasion and placenta development, and a high ratio of embryo loss are observed in Enpp2±, lpar1-/- or PPARG-blocked pregnant mice. Patients with unexplained spontaneous abortion display insufficient autophagy and cell residence of dMφ. In therapeutic studies, supplementation with LPA or the autophagy inducer rapamycin significantly promotes dMφ autophagy and cell residence, and improves embryo resorption in Enpp2± and spontaneous abortion mouse models, which should be dependent on the activation of DDIT4-autophagy-CLDN7-adhesion molecules axis. This observation reveals that inactivation of ENPP2-LPA metabolism and insufficient autophagy of dMφ result in resident obstacle of dMφ and further increase the risk of spontaneous abortion, and provides potential therapeutic strategies to prevent spontaneous abortion.Abbreviations: ACTB: actin beta; ADGRE1/F4/80: adhesion G protein-coupled receptor E1; Atg5: autophagy related 5; ATG13: autophagy related 13; BECN1: beclin 1; CDH1/E-cadherin: cadherin 1; CDH5/VE-cadherin: cadherin 5; CFSE: carboxyfluorescein succinimidyl ester; CLDN7: claudin 7; CSF1/M-CSF: colony stimulating factor 1; CSF2/GM-CSF: colony stimulating factor 2; Ctrl: control; CXCL10/IP-10: chemokine (C-X-C) ligand 10; DDIT4: DNA damage inducible transcript 4; dMφ: decidual macrophage; DSC: decidual stromal cells; ENPP2/ATX: ectonucleotide pyrophosphatase/phosphodiesterase 2; Enpp2±: Enpp2 heterozygous knockout mouse; ENPP2i/PF-8380: ENPP2 inhibitor; EPCAM: epithelial cell adhesion molecule; ESC: endometrial stromal cells; FGF2/b-FGF: fibroblast growth factor 2; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GPCPD1: glycerophosphocholine phosphodiesterase 1; HE: heterozygote; HIF1A: hypoxia inducible factor 1 subunit alpha; HNF4A: hepatocyte nuclear factor 4 alpha; HO: homozygote; ICAM2: intercellular adhesion molecule 2; IL: interleukin; ITGAV/CD51: integrin subunit alpha V; ITGAM/CD11b: integrin subunit alpha M; ITGAX/CD11b: integrin subunit alpha X; ITGB3/CD61: integrin subunit beta 3; KLRB1/NK1.1: killer cell lectin like receptor B1; KRT7/cytokeratin 7: keratin 7; LPA: lysophosphatidic acid; LPAR: lysophosphatidic acid receptor; lpar1-/-: lpar1 homozygous knockout mouse; LPAR1i/AM966: LPAR1 inhibitor; LY6C: lymphocyte antigen 6 complex, locus C1; LYPLA1: lysophospholipase 1; LYPLA2: lysophospholipase 2; Lyz2: lysozyme 2; MAP1LC3B: microtubule associated protein 1 light chain 3 beta; MARVELD2: MARVEL domain containing 2; 3-MA: 3-methyladenine; MBOAT2: membrane bound O-acyltransferase domain containing 2; MGLL: monoglyceride lipase; MRC1/CD206: mannose receptor C-type 1; MTOR: mechanistic target of rapamycin kinase; NP: normal pregnancy; PDGF: platelet derived growth factor; PLA1A: phospholipase A1 member A; PLA2G4A: phospholipase A2 group IVA; PLPP1: phospholipid phosphatase 1; pMo: peripheral blood monocytes; p-MTOR: phosphorylated MTOR; PPAR: peroxisome proliferator activated receptor; PPARG/PPARγ: peroxisome proliferator activated receptor gamma; PPARGi/GW9662: PPARG inhibitor; PTPRC/CD45: protein tyrosine phosphatase receptor type, C; Rapa: rapamycin; RHEB: Ras homolog, mTORC1 binding; SA: spontaneous abortion; SELE: selectin E; SELL: selectin L; siCLDN7: CLDN7-silenced; STAT: signal transducer and activator of transcription; SQSTM1: sequestosome 1; TJP1: tight junction protein 1; VCAM1: vascular cell adhesion molecule 1; WT: wild type.
Keywords: Abortion; CLDN7; DDIT4; ENPP2; LPAR1; decidual macrophage; lysophosphatidic acid; trophoblast invasion.
Conflict of interest statement
No potential conflict of interest was reported by the author(s)
Figures







References
-
- Garrido-Gimenez C, Alijotas-Reig J.. Recurrent miscarriage: causes, evaluation and management. Postgrad Med J. 2015;91(1073):151–162. - PubMed
-
- Alijotas-Reig J, Garrido-Gimenez C. Current concepts and new trends in the diagnosis and management of recurrent miscarriage. Obstet Gynecol Surv. 2013;68(6):445–466. - PubMed
-
- Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. 2013;31(1):387–411. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
