Autophagy deficiency abolishes liver mitochondrial DNA segregation
- PMID: 35220898
- PMCID: PMC9542960
- DOI: 10.1080/15548627.2022.2038501
Autophagy deficiency abolishes liver mitochondrial DNA segregation
Abstract
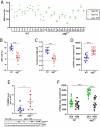
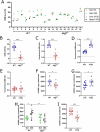
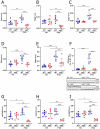
Mutations in the mitochondrial genome (mtDNA) are ubiquitous in humans and can lead to a broad spectrum of disorders. However, due to the presence of multiple mtDNA molecules in the cell, co-existence of mutant and wild-type mtDNAs (termed heteroplasmy) can mask disease phenotype unless a threshold of mutant molecules is reached. Importantly, the mutant mtDNA level can change across lifespan as mtDNA segregates in an allele- and cell-specific fashion, potentially leading to disease. Segregation of mtDNA is mainly evident in hepatic cells, resulting in an age-dependent increase of mtDNA variants, including non-synonymous potentially deleterious mutations. Here we modeled mtDNA segregation using a well-established heteroplasmic mouse line with mtDNA of NZB/BINJ and C57BL/6N origin on a C57BL/6N nuclear background. This mouse line showed a pronounced age-dependent NZB mtDNA accumulation in the liver, thus leading to enhanced respiration capacity per mtDNA molecule. Remarkably, liver-specific atg7 (autophagy related 7) knockout abolished NZB mtDNA accumulat ion, resulting in close-to-neutral mtDNA segregation through development into adulthood. prkn (parkin RBR E3 ubiquitin protein ligase) knockout also partially prevented NZB mtDNA accumulation in the liver, but to a lesser extent. Hence, we propose that age-related liver mtDNA segregation is a consequence of macroautophagic clearance of the less-fit mtDNA. Considering that NZB/BINJ and C57BL/6N mtDNAs have a level of divergence comparable to that between human Eurasian and African mtDNAs, these findings have potential implications for humans, including the safe use of mitochondrial replacement therapy.Abbreviations: Apob: apolipoprotein B; Atg1: autophagy-related 1; Atg7: autophagy related 7; Atp5a1: ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1; BL6: C57BL/6N mouse strain; BNIP3: BCL2/adenovirus E1B interacting protein 3; FCCP: carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; MAP1LC3A: microtubule-associated protein 1 light chain 3 alpha; MAP1LC3B: microtubule-associated protein 1 light chain 3 beta; mt-Atp8: mitochondrially encoded ATP synthase 8; MT-CO1: mitochondrially encoded cytochrome c oxidase I; MT-CO2: mitochondrially encoded cytochrome c oxidase II; mt-Co3: mitochondrially encoded cytochrome c oxidase III; mt-Cytb: mitochondrially encoded cytochrome b; mtDNA: mitochondrial DNA; MUL1: mitochondrial ubiquitin ligase activator of NFKB 1; nDNA: nuclear DNA; Ndufa9: NADH:ubiquinone oxireductase subunit A9; NDUFB8: NADH:ubiquinone oxireductase subunit B8; Nnt: nicotinamide nucleotide transhydrogenase; NZB: NZB/BINJ mouse strain; OXPHOS: oxidative phosphorylation; PINK1: PTEN induced putative kinase 1; Polg2: polymerase (DNA directed), gamma 2, accessory subunit; Ppara: peroxisome proliferator activated receptor alpha; Ppia: peptidylprolyl isomerase A; Prkn: parkin RBR E3 ubiquitin protein ligase; P10: post-natal day 10; P21: post-natal day 21; P100: post-natal day 100; qPCR: quantitative polymerase chain reaction; Rpl19: ribosomal protein L19; Rps18: ribosomal protein S18; SD: standard deviation; SEM: standard error of the mean; SDHB: succinate dehydrogenase complex, subunit B, iron sulfur (Ip); SQSTM1: sequestosome 1; Ssbp1: single-stranded DNA binding protein 1; TFAM: transcription factor A, mitochondrial; Tfb1m: transcription factor B1, mitochondrial; Tfb2m: transcription factor B2, mitochondrial; TOMM20: translocase of outer mitochondrial membrane 20; UQCRC2: ubiquinol cytochrome c reductase core protein 2; WT: wild-type.
Keywords: Atg7; NZB; heteroplasmy; mitochondria; mitophagy; parkin.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Craven L, Alston CL, Taylor RW, et al. Recent advances in mitochondrial disease. Annu Rev Genomics Hum Genet. 2017;18(1):257–275. - PubMed
-
- Stewart JB, Chinnery PF.. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat Rev Genet. 2020;22(2):106–118. - PubMed
-
- Wallace DC. Mitochondrial genetic medicine. Nat Genet. 2018;50(12):1642–1649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
