The Molecular Tumor Board Portal supports clinical decisions and automated reporting for precision oncology
- PMID: 35221333
- PMCID: PMC8882467
- DOI: 10.1038/s43018-022-00332-x
The Molecular Tumor Board Portal supports clinical decisions and automated reporting for precision oncology
Erratum in
-
Author Correction: The Molecular Tumor Board Portal supports clinical decisions and automated reporting for precision oncology.Nat Cancer. 2022 May;3(5):649. doi: 10.1038/s43018-022-00378-x. Nat Cancer. 2022. PMID: 35449310 Free PMC article. No abstract available.
Abstract
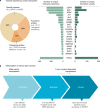
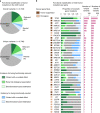
There is a growing need for systems that efficiently support the work of medical teams at the precision-oncology point of care. Here, we present the implementation of the Molecular Tumor Board Portal (MTBP), an academic clinical decision support system developed under the umbrella of Cancer Core Europe that creates a unified legal, scientific and technological platform to share and harness next-generation sequencing data. Automating the interpretation and reporting of sequencing results decrease the need for time-consuming manual procedures that are prone to errors. The adoption of an expert-agreed process to systematically link tumor molecular profiles with clinical actions promotes consistent decision-making and structured data capture across the connected centers. The use of information-rich patient reports with interactive content facilitates collaborative discussion of complex cases during virtual molecular tumor board meetings. Overall, streamlined digital systems like the MTBP are crucial to better address the challenges brought by precision oncology and accelerate the use of emerging biomarkers.
© 2022. The Author(s).
Conflict of interest statement
D.T. reports receiving honoraria for speaker activities and advisory role for Roche. R.D. reports receiving honoraria for speaker activities from Roche, Ipsen, Amgen, Sanofi, Servier Laboratories, Merck and Sharp & Dohme; an advisory role at Roche and Boehringer Ingelheim; and research grants from Merck and Pierre Fabre. I.B. reports consultant or advisory role for Orion Pharma; speaker activities for BMS; and travel grants from AstraZeneca and Merck Serono and is principal investigator of clinical trials for AstraZeneca, BMS, Celgene, Gliknik, GSK, Janssen, KURA, MSD, Novartis, Orion Pharma, Pfizer, Shattuck, Northern Biologics, Rakutan Aspirian and Nanobiotics. R.B. reports consultant or advisory roles (with funding to institution) for AstraZeneca, Daiichi Sankyo, Lilly, Molecular Partners, Novartis, Roche and Shionogi and research grants from AstraZeneca, Boehringer Ingelheim and Genentech and is principal investigator or subinvestigator of clinical trials for Astex, AstraZeneca, Boehringer Ingelheim, Boston Therapeutics, Genentech/Roche, Johnson & Johnson, Lilly, Molecular Partners, PharmaMar, Roche, Sanofi-Aventis, Shionogi and Taiho. Y.L. has received honoraria for participation in advisory boards for Merck KGaA, Pfizer, Gilead/Immunomedics, Seattle Genetics and Taiho Pharma; lecture support from MSD, AstraZeneca, Astellas, Janssen, Roche and BMS; institutional research grants from Taiho, Sanofi, MSD and Celsius; institutional research support as a local principal investigator from Pfizer, Janssen, Exelexis, AstraZeneca, Pfizer, Merck KGaA, BMS, Astellas, Gilead and Incyte; and steering committee membership for Astellas, Gilead/Immunomedics, Basilea Pharmaceutica and Taiho. C.M. reports consultant/advisory fees from Amgen, Astellas, AstraZeneca, Bayer, BeiGene, BMS, Celgene, Debiopharm, Genentech, Ipsen, Janssen, Lilly, MedImmune, MSD, Novartis, Pfizer, Roche, Sanofi and Orion and is a principal investigator/subinvestigator of clinical trials for AbbVie, Aduro, Agios, Amgen, Argen-x, Astex, AstraZeneca, Aveo Pharmaceuticals, Bayer, BeiGene, Blueprint, BMS, Boehringer Ingelheim, Celgene, Chugai, Clovis, Daiichi Sankyo, Debiopharm, Eisai, Eos, Exelixis, Forma, Gamamabs, Genentech, Gortec, GSK, H3 Biomedicine, Incyte, Innate Pharma, Janssen, Kura Oncology, Kyowa, Lilly, Loxo, Lysarc, Lytix Biopharma, Medimmune, Menarini, Merus, MSD, Nanobiotix, Nektar Therapeutics, Novartis, Octimet, Oncoethix, Oncopeptides AB, Orion, Pfizer, PharmaMar, Pierre Fabre, Roche, Sanofi, Servier, Sierra Oncology, Taiho, Takeda, Tesaro and Xencor. R.S. reports research funding from Pfizer, AstraZeneca, PharmaMar, Roche and Daiichi Sankyo; speakers honoraria from Pfizer, Daiichi Sankyo and Novartis; and participation in ad boards for Pfizer, Daiichi Sankyo and Novartis. C.V. reports receiving honoraria for advisory role for Novartis. G.A. reports no conflicts of interests with regards to the topic at hand; as scientific director of the Fondazione IRCCS Istituto Nazionale dei Tumori, he is legally responsible for contracts with Pharma and other funding agencies. C.C. is a member of the External Science Panel of AstraZeneca and Illumina’s Scientific Advisory Board, and his laboratory has received research grants (administered by the University of Cambridge) from Genentech, Roche, AstraZeneca and Servier. S.F. reports a consulting or advisory role, having received honoraria, research funding and/or travel/accommodation expenses funding from the following for-profit companies: Bayer, Roche, Amgen, Eli Lilly, PharmaMar, AstraZeneca and Pfizer. E.G. reports consultant honoraria from Roche/Genentech, F. Hoffmann-La Roche, Ellipses Pharma, Neomed Therapeutics, Boehringer Ingelheim-Janssen Global Services, AstraZeneca, SeaGen and TFS-Alkermes; research funding from Novartis/Roche; is a principal investigator/subinvestigator of clinical trials for Principia Biopharma, Lilly, Novartis Farmacéutica, Genentech, Loxo Oncology, F. Hoffmann-La Roche, Symphogen A/S, Merck, Sharp & Dohme de España, Incyte Biosciences International, PharmaMar, Kura Oncology, Macrogenics, Glycotope, Pierre Fabre Medicament, Cellestia Biotech, Menarini Ricerche Spa, Blueprint Medicines Corporation, BeiGene, Sierra Oncology and Genmab B.V; travel grants from Bristol Myers Squibb, Merck Sharp & Dohme, Menarini and Glycotope; and is on the speakers bureau for Bristol Myers Squibb, Merck Sharp & Dohme, Roche and ThermoFisher. J.T. reports personal financial interests in the form of a scientific consultancy role for Array Biopharma, AstraZeneca, Bayer, BeiGene, Boehringer Ingelheim, Chugai, Genentech, Genmab A/S, Halozyme, Imugene, Inflection Biosciences, Ipsen, Kura Oncology, Lilly, MSD, Menarini, Merck Serono, Merrimack, Merus, Molecular Partners, Novartis, Peptomyc, Pfizer, Pharmacyclics, ProteoDesign SL, Rafael Pharmaceuticals, F. Hoffmann-La Roche, Sanofi, SeaGen, Seattle Genetics, Servier, Symphogen, Taiho, VCN Biosciences, Biocartis, Foundation Medicine, HalioDX SAS and Roche Diagnostics. E.V. reports no conflict of interest with regards to the topic at hand; as medical director of the Netherlands Cancer Institute, he is legally responsible for all contracts with pharma. J.R. reports research funding from Bayer & Novartis; clinical research for Spectrum Pharmaceuticals, Tocagen, Symphogen, BioAlta, Pfizer, GenMab, CytomX, KELUN Biotech, Takeda-Millennium, GlaxoSmithKline and IPSEN; scientific advisory board membership for Novartis, Eli Lilly, Orion Pharmaceuticals, Servier Pharmaceuticals, Peptomyc, Merck Sharp & Dohme, Kelun Pharmaceuticals/Klus Pharma, Spectrum Pharmaceuticals, Pfizer, Roche Pharmaceuticals and Ellipses Pharma; and research funding from Bayer & Novartis. J.L. reports research funding from AstraZeneca, Novartis and GE Healthcare and is cofounder and shareholder of FenoMark Diagnostics. The remaining authors declare no competing interests.
Figures