Radiation exposure elicits a neutrophil-driven response in healthy lung tissue that enhances metastatic colonization
- PMID: 35221334
- PMCID: PMC7612918
- DOI: 10.1038/s43018-022-00336-7
Radiation exposure elicits a neutrophil-driven response in healthy lung tissue that enhances metastatic colonization
Erratum in
-
Author Correction: Radiation exposure elicits a neutrophil-driven response in healthy lung tissue that enhances metastatic colonization.Nat Cancer. 2022 Apr;3(4):519. doi: 10.1038/s43018-022-00373-2. Nat Cancer. 2022. PMID: 35379986 No abstract available.
Abstract
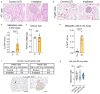
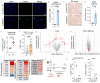
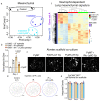
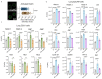
Radiotherapy is one of the most effective approaches to achieve tumor control in cancer patients, although healthy tissue injury due to off-target radiation exposure can occur. In this study, we used a model of acute radiation injury to the lung, in the context of cancer metastasis, to understand the biological link between tissue damage and cancer progression. We exposed healthy mouse lung tissue to radiation before the induction of metastasis and observed a strong enhancement of cancer cell growth. We found that locally activated neutrophils were key drivers of the tumor-supportive preconditioning of the lung microenvironment, governed by enhanced regenerative Notch signaling. Importantly, these tissue perturbations endowed arriving cancer cells with an augmented stemness phenotype. By preventing neutrophil-dependent Notch activation, via blocking degranulation, we were able to significantly offset the radiation-enhanced metastases. This work highlights a pro-tumorigenic activity of neutrophils, which is likely linked to their tissue regenerative functions.
© 2022. Crown.
Conflict of interest statement
The authors’ declare no competing interests.
Figures





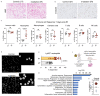




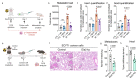

Comment in
-
Radiation sparks a dark side in neutrophils.Nat Cancer. 2022 Feb;3(2):136-137. doi: 10.1038/s43018-021-00324-3. Nat Cancer. 2022. PMID: 35221335 No abstract available.
-
Naughty neutrophils.Nat Rev Cancer. 2022 May;22(5):258. doi: 10.1038/s41568-022-00470-5. Nat Rev Cancer. 2022. PMID: 35314809 No abstract available.
References
-
- Vicini FA, Kestin L, Huang R, Martinez A. Does local recurrence affect the rate of distant metastases and survival in patients with early-stage breast carcinoma treated with breast-conserving therapy? Cancer. 2003;97:910–919. - PubMed
-
- Kuonen F, Secondini C, Rüegg C. Molecular pathways: emerging pathways mediating growth, invasion, and metastasis of tumors progressing in an irradiated microenvironment. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:5196–5202. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

