A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology
- PMID: 35221336
- PMCID: PMC8882468
- DOI: 10.1038/s43018-022-00337-6
A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology
Abstract
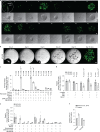
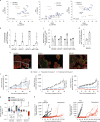
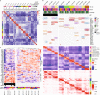
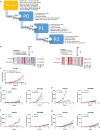
Models that recapitulate the complexity of human tumors are urgently needed to develop more effective cancer therapies. We report a bank of human patient-derived xenografts (PDXs) and matched organoid cultures from tumors that represent the greatest unmet need: endocrine-resistant, treatment-refractory and metastatic breast cancers. We leverage matched PDXs and PDX-derived organoids (PDxO) for drug screening that is feasible and cost-effective with in vivo validation. Moreover, we demonstrate the feasibility of using these models for precision oncology in real time with clinical care in a case of triple-negative breast cancer (TNBC) with early metastatic recurrence. Our results uncovered a Food and Drug Administration (FDA)-approved drug with high efficacy against the models. Treatment with this therapy resulted in a complete response for the individual and a progression-free survival (PFS) period more than three times longer than their previous therapies. This work provides valuable methods and resources for functional precision medicine and drug development for human breast cancer.
© 2022. The Author(s).
Conflict of interest statement
University of Utah may license the models described herein to for-profit companies, which may result in tangible property royalties to members of the Welm lab who developed the models (K.P.G., M.F., A.J.B., S.D.S., Z.C., Y.S.D., L.Z., E.C.-S., C.-H.Y., J.T., G.W., A.L.W. and B.E.W.). M.T.L. is a Manager in StemMed Holdings LLC, a limited partner in StemMed Ltd, and holds an equity stake in Tvardi Therapeutics. L.E.D. is a compensated employee of StemMed, Ltd. S.O. has received research support/reagents from AstraZeneca, Illumina, H3 Biomedicine and Blueprint Medicine. The other authors declare no conflicts.
Figures










Comment in
-
A new sophistication for breast cancer PDXs.Nat Cancer. 2022 Feb;3(2):138-140. doi: 10.1038/s43018-021-00328-z. Nat Cancer. 2022. PMID: 35221337 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

