Differential Gene Sets Profiling in Gram-Negative and Gram-Positive Sepsis
- PMID: 35223539
- PMCID: PMC8863667
- DOI: 10.3389/fcimb.2022.801232
Differential Gene Sets Profiling in Gram-Negative and Gram-Positive Sepsis
Abstract
Background: The host response to bacterial sepsis is reported to be nonspecific regardless of the causative pathogen. However, newer paradigms indicated that the host response of Gram-negative sepsis may be different from Gram-positive sepsis, and the difference has not been clearly clarified. The current study aimed to explore the difference by identifying the differential gene sets using the genome-wide technique.
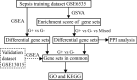
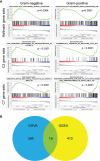
Methods: The training dataset GSE6535 and the validation dataset GSE13015 were used for bioinformatics analysis. The distinct gene sets of sepsis with different infections were screened using gene set variation analysis (GSVA) and gene set enrichment analysis (GSEA). The intersection gene sets based on the two algorithms were confirmed through Venn analysis. Finally, the common gene sets between GSE6535 and GSE13015 were determined by GSEA.
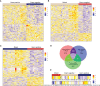
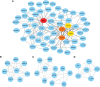
Results: Two immunological gene sets in GSE6535 were identified based on GSVA, which could be used to discriminate sepsis caused by Gram-positive, Gram-negative, or mixed infection. A total of 19 gene sets were obtained in GSE6535 through Venn analysis based on GSVA and GSEA, which revealed the heterogeneity of Gram-negative and Gram-positive sepsis at the molecular level. The result was also verified by analysis of the validation set GSE13015, and 40 common differential gene sets were identified between dataset GSE13015 and dataset GSE6535 by GSEA.
Conclusions: The identified differential gene sets indicated that host response may differ dramatically depending on the inciting organism. The findings offer new insight to investigate the pathophysiology of bacterial sepsis.
Keywords: Gram-negative; Gram-positive; gene sets; microarray analysis; sepsis.
Copyright © 2022 Wang, Li, Tang, Guan, Xiong, Zhu, Gong and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria.Infect Immun. 2003 Oct;71(10):5803-13. doi: 10.1128/IAI.71.10.5803-5813.2003. Infect Immun. 2003. PMID: 14500502 Free PMC article.
-
Differential gene expression in gram-negative and gram-positive sepsis.Am J Respir Crit Care Med. 2004 May 15;169(10):1135-43. doi: 10.1164/rccm.200211-1278OC. Epub 2004 Mar 4. Am J Respir Crit Care Med. 2004. PMID: 15001460
-
Transcriptomic data from two primary cell models stimulating human monocytes suggest inhibition of oxidative phosphorylation and mitochondrial function by N. meningitidis which is partially up-regulated by IL-10.BMC Immunol. 2017 Oct 27;18(1):46. doi: 10.1186/s12865-017-0229-5. BMC Immunol. 2017. PMID: 29078758 Free PMC article.
-
Clinical gram-positive sepsis: does it fundamentally differ from gram-negative bacterial sepsis?Crit Care Med. 1999 Aug;27(8):1608-16. doi: 10.1097/00003246-199908000-00039. Crit Care Med. 1999. PMID: 10470773 Review.
-
Host gene expression profiling in pathogen-host interactions.Curr Opin Immunol. 2006 Aug;18(4):422-9. doi: 10.1016/j.coi.2006.05.018. Epub 2006 Jun 19. Curr Opin Immunol. 2006. PMID: 16782318 Review.
Cited by
-
Changes of Serum Pyruvate Kinase M2 Level in Patients with Sepsis and Its Clinical Value.Infect Drug Resist. 2023 Sep 28;16:6437-6449. doi: 10.2147/IDR.S429314. eCollection 2023. Infect Drug Resist. 2023. PMID: 37795205 Free PMC article.
-
Effects of Sepsis on Immune Response, Microbiome and Oxidative Metabolism in Preterm Infants.Children (Basel). 2023 Mar 22;10(3):602. doi: 10.3390/children10030602. Children (Basel). 2023. PMID: 36980160 Free PMC article. Review.
-
Differential neutrophil responses in murine following intraperitoneal injections of Escherichia coli and Staphylococcus aureus.Heliyon. 2024 Nov 15;10(22):e40281. doi: 10.1016/j.heliyon.2024.e40281. eCollection 2024 Nov 30. Heliyon. 2024. PMID: 39641065 Free PMC article.
-
Transcriptional markers classifying Escherichia coli and Staphylococcus aureus induced sepsis in adults: A data-driven approach.PLoS One. 2024 Jul 5;19(7):e0305920. doi: 10.1371/journal.pone.0305920. eCollection 2024. PLoS One. 2024. PMID: 38968271 Free PMC article.
-
Antibiotic-mediated dysbiosis leads to activation of inflammatory pathways.Front Immunol. 2025 Jan 9;15:1493991. doi: 10.3389/fimmu.2024.1493991. eCollection 2024. Front Immunol. 2025. PMID: 39850904 Free PMC article.
References
-
- Carson W., Salter-Green S. E., Scola M. M., Joshi A., Gallagher K. A., Kunkel S. L. (2017). Enhancement of Macrophage Inflammatory Responses by CCL2 is Correlated With Increased miR-9 Expression and Downregulation of the ERK1/2 Phosphatase Dusp6. Cell Immunol. 314, 63–72. doi: 10.1016/j.cellimm.2017.02.005 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

