Transcriptional Regulation of the Creatine Utilization Genes of Corynebacterium glutamicum ATCC 14067 by AmtR, a Central Nitrogen Regulator
- PMID: 35223787
- PMCID: PMC8864220
- DOI: 10.3389/fbioe.2022.816628
Transcriptional Regulation of the Creatine Utilization Genes of Corynebacterium glutamicum ATCC 14067 by AmtR, a Central Nitrogen Regulator
Abstract
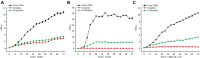
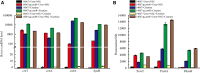
In the genus Corynebacterium, AmtR is a key component of the nitrogen regulatory system, and it belongs to the TetR family of transcription regulators. There has been much research on AmtR structure, functions, and regulons in the type strain C. glutamicum ATCC 13032, but little research in other C. glutamicum strains. In this study, chromatin immunoprecipitation and massively parallel DNA sequencing (ChIP-seq) was performed to identify the AmtR regulon in C. glutamicum ATCC 14067. Ten peaks were obtained in the C. glutamicum ATCC 14067 genome including two new peaks related to three operons (RS_01910-RS_01915, RS_15995, and RS_16000). The interactions between AmtR and the promoter regions of the three operons were confirmed by electrophoretic mobility shift assays (EMSAs). The RS_01910, RS_01915, RS_15995, and RS_16000 are not present in the type strain C. glutamicum ATCC 13032. Sequence analysis indicates that RS_01910, RS_01915, RS_15995, and RS_16000, are related to the degradation of creatine and creatinine; RS_01910 may encode a protein related to creatine transport. The genes RS_01910, RS_01915, RS_15995, and RS_16000 were given the names crnA, creT, cshA, and hyuB, respectively. Real-time quantitative PCR (RT-qPCR) analysis and sfGFP (superfolder green fluorescent protein) analysis reveal that AmtR directly and negatively regulates the transcription and expression of crnA, creT, cshA, and hyuB. A growth test shows that C. glutamicum ATCC 14067 can use creatine or creatinine as a sole nitrogen source. In comparison, a creT deletion mutant strain is able to grow on creatinine but loses the ability to grow on creatine. This study provides the first genome-wide captures of the dynamics of in vivo AmtR binding events and the regulatory network they define. These elements provide more options for synthetic biology by extending the scope of the AmtR regulon.
Keywords: AmtR; ChIP-seq; Corynebacterium glutamicum; MFS transporter; creatine; creatinine.
Copyright © 2022 Zhang, Ouyang, Zhao, Han and Zheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

