Development of Loop-Mediated Isothermal Amplification Rapid Diagnostic Assays for the Detection of Klebsiella pneumoniae and Carbapenemase Genes in Clinical Samples
- PMID: 35223985
- PMCID: PMC8864245
- DOI: 10.3389/fmolb.2021.794961
Development of Loop-Mediated Isothermal Amplification Rapid Diagnostic Assays for the Detection of Klebsiella pneumoniae and Carbapenemase Genes in Clinical Samples
Abstract
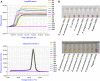
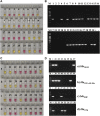
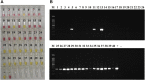
Klebsiella pneumoniae is an important pathogenic bacterium commonly associated with human healthcare and community-acquired infections. In recent years, K. pneumoniae has become a significant threat to global public and veterinary health, because of its high rates of antimicrobial resistance (AMR). Early diagnosis of K. pneumoniae infection and detection of any associated AMR would help to accelerate directed therapy and reduce the risk of the emergence of multidrug-resistant isolates. In this study, we identified three target genes (yhaI, epsL, and xcpW) common to K. pneumoniae isolates from both China and Europe and designed loop-mediated isothermal amplification (LAMP) assays for the detection of K. pneumoniae in clinical samples. We also designed LAMP assays for the detection of five AMR genes commonly associated with K. pneumoniae. The LAMP assays were validated on a total of 319 type reference strains and clinical isolates of diverse genetic backgrounds, in addition to 40 clinical human sputum samples, and were shown to be reliable, highly specific, and sensitive. For the K. pneumoniae-specific LAMP assay, the calculated sensitivity, specificity, and positive and negative predictive values (comparison with culture and matrix-assisted laser desorption/ionization-time of flight mass spectrometry) were all 100% on clinical isolates and, respectively, of 100%, 91%, and 90%, and 100% when tested on clinical sputum samples, while being significantly faster than the reference methods. For the bla KPC and other carbapenemases' LAMP assays, the concordance between the LAMP results and the references methods (susceptibility tests) was 100%, on both pure cultures (n = 125) and clinical samples (n = 18). In conclusion, we developed highly sensitive and specific LAMP assays for the clinical identification of K. pneumoniae and detection of carbapenem resistance.
Keywords: Klebsiella pneumoniae (K. pneumoniae); LAMP (loop mediated isothermal amplification); antimicrobial resistance (AMR); carbapenemases genes; rapid diagnostics.
Copyright © 2022 Poirier, Kuang, Siedler, Borah, Mehat, Liu, Tai, Wang, van Vliet, Ma, Jenkins, Clark, La Ragione, Qu and McFadden.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Adeolu M., Alnajar S., Naushad S., S. Gupta R. (2016). Genome-based Phylogeny and Taxonomy of the 'Enterobacteriales': Proposal for Enterobacterales ord. Nov. Divided into the Families Enterobacteriaceae, Erwiniaceae Fam. nov., Pectobacteriaceae Fam. nov., Yersiniaceae Fam. nov., Hafniaceae Fam. nov., Morganellaceae Fam. nov., and Budviciaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 66 (12), 5575–5599. 10.1099/ijsem.0.001485 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

