Predicting Diagnostic Gene Biomarkers Associated With Immune Checkpoints, N6-Methyladenosine, and Ferroptosis in Patients With Acute Myocardial Infarction
- PMID: 35224064
- PMCID: PMC8873927
- DOI: 10.3389/fcvm.2022.836067
Predicting Diagnostic Gene Biomarkers Associated With Immune Checkpoints, N6-Methyladenosine, and Ferroptosis in Patients With Acute Myocardial Infarction
Abstract
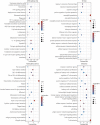
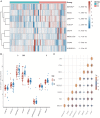
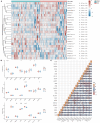
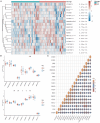
This study aimed to determine early diagnosis genes of acute myocardial infarction (AMI) and then validate their association with ferroptosis, immune checkpoints, and N6-methyladenosine (m6A), which may provide a potential method for the early diagnosis of AMI. Firstly, we downloaded microarray data from NCBI (GSE61144, GSE60993, and GSE42148) and identified differentially expressed genes (DEGs) in samples from healthy subjects and patients with AMI. Also, we performed systematic gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses and used STRING to predict protein interactions. Moreover, MCC and MCODE algorithms in the cytoHubba plug-in were used to screen nine key genes in the network. We then determined the diagnostic significance of the nine obtained DEGs by plotting receiver operating characteristic curves using a multiscale curvature classification algorithm. Meanwhile, we investigated the relationship between AMI and immune checkpoints, ferroptosis, and m6A. In addition, we further validated the key genes through the GSE66360 dataset and consequently obtained nine specific genes that can be used as early diagnosis biomarkers for AMI. Through screening, we identified 210 DEGs, including 53 downregulated and 157 upregulated genes. According to GO, KEGG, and key gene screening results, FPR1, CXCR1, ELANE, TLR2, S100A12, TLR4, CXCL8, FPR2 and CAMP could be used for early prediction of AMI. Finally, we found that AMI was associated with ferroptosis, immune checkpoints, and m6A and FPR1, CXCR1, ELANE, TLR2, S100A12, TLR4, CXCL8, FPR2 and CAMP are effective markers for the diagnosis of AMI, which can provide new prospects for future studies on the pathogenesis of AMI.
Keywords: acute myocardial infarction; diagnostic gene biomarker; differentially expressed genes; ferroptosis; immune checkpoints; m6A.
Copyright © 2022 Tong, Zhao, Dang, Kou and Kou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

