Tumor microenvironment pathways: Cross regulation in breast cancer metastasis
- PMID: 35224148
- PMCID: PMC8843880
- DOI: 10.1016/j.gendis.2020.11.015
Tumor microenvironment pathways: Cross regulation in breast cancer metastasis
Abstract
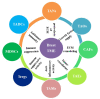
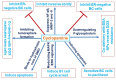
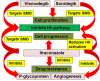
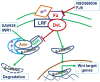
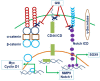
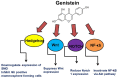
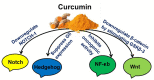
The tumor microenvironment (TME) is heterogeneous and contains a multiple cell population with surrounded immune cells, which plays a major role in regulating metastasis. The multifunctional pathways, Hedgehog (Hh), Wnt, Notch, and NF-kB, cross-regulates metastasis in breast cancer. This review presents substantial evidence for cross-regulation of TME components and signaling pathways, which makes breast TME more heterogeneous and complex, promoting breast cancer progression and metastasis as a highly aggressive form. We discoursed the importance of stromal and immune cells as well as their crosstalk in bridging the metastasis. We also discussed the role of Hh and Notch pathways in the intervention between breast cancer cells and macrophages to support TME; Notch signaling in the bidirectional communication between cancer cells and components of TME; Wnt signal pathway in controlling the factors responsible for EMT and NF-κB pathway in the regulation of genes controlling the inflammatory response. We also present the role of exosomes and their miRNAs in the cross-regulation of TME cells as well as pathways in the reprogramming of breast TME to support metastasis. Finally, we examined and discussed the targeted small molecule inhibitors and natural compounds targeting developmental pathways and proposed small molecule natural compounds as potential therapeutics of TME based on the multitargeting ability. In conclusion, the understanding of the molecular basis of the cross-regulation of TME pathways and their inhibitors helps identify molecular targets for rational drug discovery to treat breast cancers.
Keywords: Breast cancer; Cross-regulation; Hedgehog; NF-κB; Notch; Tumor microenvironment; Wnt.
© 2020 Chongqing Medical University. Production and hosting by Elsevier B.V.
Figures

References
-
- Key T.J., Verkasalo P.K., Banks E. Epidemiology of breast cancer. Lancet Oncol. 2001;2(3):133–140. - PubMed
-
- Ingham P.W., McMahon A.P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–3087. - PubMed
-
- Barth S.D., Schulze J.J., Kühn T., et al. Treg-mediated immune tolerance and the risk of solid cancers: findings from EPIC-Heidelberg. J Natl Cancer Inst. 2015;107(11):djv224. - PubMed
-
- Linke F., Harenberg M., Nietert M.M., et al. Microenvironmental interactions between endothelial and lymphoma cells: a role for the canonical WNT pathway in Hodgkin lymphoma. Leukemia. 2017;31(2):361–372. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

