Cystine Knot Peptides with Tuneable Activity and Mechanism
- PMID: 35224831
- PMCID: PMC9539897
- DOI: 10.1002/anie.202200951
Cystine Knot Peptides with Tuneable Activity and Mechanism
Abstract
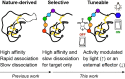
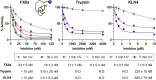
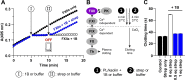
Knottins are topologically complex peptides that are stabilised by a cystine knot and have exceptionally diverse functions, including protease inhibition. However, approaches for tuning their activity in situ are limited. Here, we demonstrate separate approaches for tuning the activity of knottin protease inhibitors using light or streptavidin. We show that the inhibitory activity and selectivity of an engineered knottin can be controlled with light by activating a second mode of action that switches the inhibitor ON against new targets. Guided by a knottin library screen, we also identify a position in the inhibitor's binding loop that permits insertion of a biotin tag without impairing activity. Using streptavidin, biotinylated knottins with nanomolar affinity can be switched OFF in activity assays, and the anticoagulant activity of a factor XIIa inhibitor can be rapidly switched OFF in human plasma. Our findings expand the scope of engineered knottins for precisely controlling protein function.
Keywords: Activity Switch; Enzymes; Inhibitors; Knottins; Photoactivation.
© 2022 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Cystine-knot peptides: emerging tools for cancer imaging and therapy.Expert Rev Proteomics. 2014 Oct;11(5):561-72. doi: 10.1586/14789450.2014.932251. Epub 2014 Aug 28. Expert Rev Proteomics. 2014. PMID: 25163524 Review.
-
High-Affinity RGD-Knottin Peptide as a New Tool for Rapid Evaluation of the Binding Strength of Unlabeled RGD-Peptides to αvβ3, αvβ5, and α5β1 Integrin Receptors.Anal Chem. 2017 Jun 6;89(11):5991-5997. doi: 10.1021/acs.analchem.7b00554. Epub 2017 May 24. Anal Chem. 2017. PMID: 28492301
-
Engineered knottin peptides as diagnostics, therapeutics, and drug delivery vehicles.Curr Opin Chem Biol. 2016 Oct;34:143-150. doi: 10.1016/j.cbpa.2016.08.022. Epub 2016 Sep 16. Curr Opin Chem Biol. 2016. PMID: 27642714 Review.
-
Engineering agatoxin, a cystine-knot peptide from spider venom, as a molecular probe for in vivo tumor imaging.PLoS One. 2013;8(4):e60498. doi: 10.1371/journal.pone.0060498. Epub 2013 Apr 3. PLoS One. 2013. PMID: 23573262 Free PMC article.
-
Biological diversity and therapeutic potential of natural and engineered cystine knot miniproteins.Curr Opin Pharmacol. 2009 Oct;9(5):608-14. doi: 10.1016/j.coph.2009.05.004. Epub 2009 Jun 10. Curr Opin Pharmacol. 2009. PMID: 19523876 Review.
Cited by
-
Stabilizing Scaffold for Short Peptides Based on Knottins.Curr Cancer Drug Targets. 2024;24(12):1275-1285. doi: 10.2174/0115680096285288240118090050. Curr Cancer Drug Targets. 2024. PMID: 38357956
-
Disulfide-constrained peptide scaffolds enable a robust peptide-therapeutic discovery platform.PLoS One. 2024 Mar 28;19(3):e0300135. doi: 10.1371/journal.pone.0300135. eCollection 2024. PLoS One. 2024. PMID: 38547109 Free PMC article.
-
Inhibition of Staphylococcus aureus biofilm formation by gurmarin, a plant-derived cyclic peptide.Front Cell Infect Microbiol. 2022 Oct 4;12:1017545. doi: 10.3389/fcimb.2022.1017545. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36268224 Free PMC article.
-
Discovery and Characterization of MaK: A Novel Knottin Antimicrobial Peptide from Monochamus alternatus.Int J Mol Sci. 2023 Dec 17;24(24):17565. doi: 10.3390/ijms242417565. Int J Mol Sci. 2023. PMID: 38139394 Free PMC article.
-
Redox signaling-mediated tumor extracellular matrix remodeling: pleiotropic regulatory mechanisms.Cell Oncol (Dordr). 2024 Apr;47(2):429-445. doi: 10.1007/s13402-023-00884-9. Epub 2023 Oct 4. Cell Oncol (Dordr). 2024. PMID: 37792154 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

