Adenovirus 5 Vectors Expressing SARS-CoV-2 Proteins
- PMID: 35224978
- PMCID: PMC9044955
- DOI: 10.1128/msphere.00998-21
Adenovirus 5 Vectors Expressing SARS-CoV-2 Proteins
Abstract
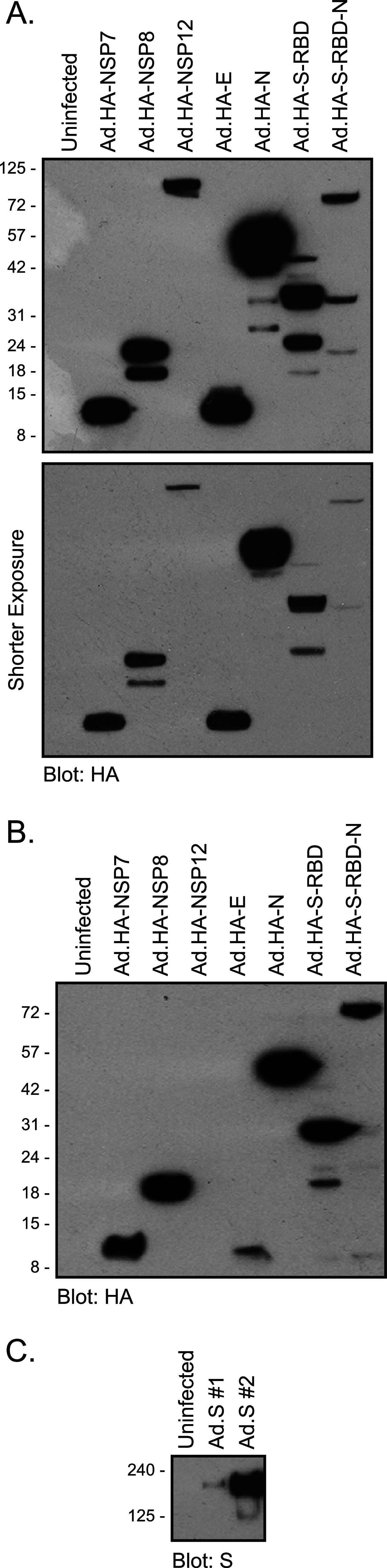
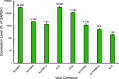
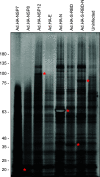
SARS-CoV-2 coronavirus is a recently identified novel coronavirus that is the causative agent of the COVID-19 pandemic that began in 2020. An intense research effort has been undertaken by the research community in order to better understand the molecular etiology of this virus and its mechanisms of host cell subjugation and immune system evasion. To facilitate further research into the SARS-CoV-2 coronavirus we have generated adenovirus 5-based viral vectors that express SARS-CoV-2 proteins-S, N, E, NSP7, NSP8, and NSP12 as hemagglutinin (HA)-tagged and untagged variants. We have also engineered two additional viruses that express the S protein receptor binding domain and a fusion of the receptor binding domain to the N protein. We show that these vectors are expressed in several different cell lines by Western blotting and real-time quantitative reverse transcriptase (qRT-PCR), we evaluate the subcellular localization of these viral proteins, and we show that these coronavirus proteins bind to a variety of cellular targets. The flexibility of adenovirus vectors allows them to be used in a variety of cell models and, importantly, in animal models as well. IMPORTANCE The COVID-19 pandemic caused by the SARS-CoV-2 coronavirus has brought untold personal and economic suffering to the world. Intense research has made tremendous progress in understanding how this virus works, yet much research remains to be done as new variants and continued evolution of the virus keep shifting the rules of engagement on the pandemic battlefield. Therefore, wide availability of resources and reagents to study SARS-CoV-2 is essential in overcoming the pandemic and for the prevention of future outbreaks. Our viral vectors provide additional tools for researchers to use in order to better understand the molecular biology of virus-host interactions and other aspects of SARS-CoV-2.
Keywords: COVID-19; SARS-CoV-2; adenovirus; viral vectors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Knipe DM, Howley PM. 2013. Fields virology, 6th ed. Wolters Kluwer/Lippincott Williams & Wilkins Health, Philadelphia, PA.
-
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for Covid-19. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi: 10.1056/NEJMoa2002032. - DOI - PMC - PubMed
-
- Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. 2020. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27:325–328. doi: 10.1016/j.chom.2020.02.001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

