Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?
- PMID: 35225672
- PMCID: PMC8941872
- DOI: 10.1128/jvi.02077-21
Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?
Abstract
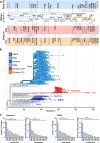
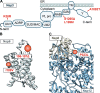
Emerging strains of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19) pandemic, that show increased transmission fitness and/or immune evasion are classified as "variants of concern" (VOCs). Recently, a SARS-CoV-2 variant first identified in November 2021 in South Africa has been recognized as a fifth VOC, termed "Omicron." What makes this VOC so alarming is the high number of changes, especially in the viral Spike protein, and accumulating evidence for increased transmission efficiency and escape from neutralizing antibodies. In an amazingly short time, the Omicron VOC has outcompeted the previously dominating Delta VOC. However, it seems that the Omicron VOC is overall less pathogenic than other SARS-CoV-2 VOCs. Here, we provide an overview of the mutations in the Omicron genome and the resulting changes in viral proteins compared to other SARS-CoV-2 strains and discuss their potential functional consequences.
Keywords: BA.1; BA.2; COVID-19; Omicron; SARS-CoV-2; Spike; variants of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- World Health Organization. 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.....
-
- Grabowski F, Kochańczyk M, Lipniacki T. 2022. The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.08.21267494v2. - DOI - PMC - PubMed
-
- Bai Y, Du Z, Xu M, Wang L, Wu P, Lau E, Cowling BJ, Meyers LA. 2021. International risk of SARS-CoV-2 Omicron variant importations originating in South Africa. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.07.21267410v1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

