PDZD8 Disruption Causes Cognitive Impairment in Humans, Mice, and Fruit Flies
- PMID: 35227461
- PMCID: PMC9302898
- DOI: 10.1016/j.biopsych.2021.12.017
PDZD8 Disruption Causes Cognitive Impairment in Humans, Mice, and Fruit Flies
Abstract
Background: The discovery of coding variants in genes that confer risk of intellectual disability (ID) is an important step toward understanding the pathophysiology of this common developmental disability.
Methods: Homozygosity mapping, whole-exome sequencing, and cosegregation analyses were used to identify gene variants responsible for syndromic ID with autistic features in two independent consanguineous families from the Arabian Peninsula. For in vivo functional studies of the implicated gene's function in cognition, Drosophila melanogaster and mice with targeted interference of the orthologous gene were used. Behavioral, electrophysiological, and structural magnetic resonance imaging analyses were conducted for phenotypic testing.
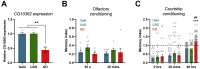
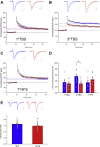
Results: Homozygous premature termination codons in PDZD8, encoding an endoplasmic reticulum-anchored lipid transfer protein, showed cosegregation with syndromic ID in both families. Drosophila melanogaster with knockdown of the PDZD8 ortholog exhibited impaired long-term courtship-based memory. Mice homozygous for a premature termination codon in Pdzd8 exhibited brain structural, hippocampal spatial memory, and synaptic plasticity deficits.
Conclusions: These data demonstrate the involvement of homozygous loss-of-function mutations in PDZD8 in a neurodevelopmental cognitive disorder. Model organisms with manipulation of the orthologous gene replicate aspects of the human phenotype and suggest plausible pathophysiological mechanisms centered on disrupted brain development and synaptic function. These findings are thus consistent with accruing evidence that synaptic defects are a common denominator of ID and other neurodevelopmental conditions.
Keywords: Brain structure; Endoplasmic reticulum; Intellectual disability; Long-term memory; PDZD8; Synaptic plasticity.
Copyright © 2022 Society of Biological Psychiatry. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- American Psychiatric Association . 5th ed. American Psychiatric Association; Washington, DC: 2013. Diagnostic and Statistical Manual of Mental Disorders.
-
- World Health Organization . World Health Organization; Geneva: 1992. The ICD-10 Classification of Mental and Behavioural Disorders: Clinical Descriptions and Diagnostic Guidelines.
-
- O’Leary C., Leonard H., Bourke J., D’Antoine H., Bartu A., Bower C. Intellectual disability: Population-based estimates of the proportion attributable to maternal alcohol use disorder during pregnancy. Dev Med Child Neurol. 2013;55:271–277. - PubMed
-
- Vissers L.E.L.M., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18. - PubMed
-
- Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W.M., Willemsen M.H., et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/R019401/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R002177/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 101029/Z/1/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MC_UP_1502/3/MRC_/Medical Research Council/United Kingdom

