Proteogenomic Analysis of Breast Cancer Transcriptomic and Proteomic Data, Using De Novo Transcript Assembly: Genome-Wide Identification of Novel Peptides and Clinical Implications
- PMID: 35227895
- PMCID: PMC9020135
- DOI: 10.1016/j.mcpro.2022.100220
Proteogenomic Analysis of Breast Cancer Transcriptomic and Proteomic Data, Using De Novo Transcript Assembly: Genome-Wide Identification of Novel Peptides and Clinical Implications
Abstract
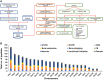
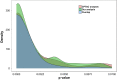
We have carried out proteogenomic analysis of the breast cancer transcriptomic and proteomic data, available at The Clinical Proteomic Tumor Analysis Consortium resource, to identify novel peptides arising from alternatively spliced events as well as other noncanonical expressions. We used a pipeline that consisted of de novo transcript assembly, six frame-translated custom database, and a combination of search engines to identify novel peptides. A portfolio of 4,387 novel peptide sequences initially identified was further screened through PepQuery validation tool (Clinical Proteomic Tumor Analysis Consortium), which yielded 1,558 novel peptides. We considered the dataset of 1,558 validated through PepQuery to understand their functional and clinical significance, leaving the rest to be further verified using other validation tools and approaches. The novel peptides mapped to the known gene sequences as well as to genomic regions yet undefined for translation, 580 novel peptides mapped to known protein-coding genes, 147 to non-protein-coding genes, and 831 belonged to novel translational sequences. The novel peptides belonging to protein-coding genes represented alternatively spliced events or 5' or 3' extensions, whereas others represented translation from pseudogenes, long noncoding RNAs, or novel peptides originating from uncharacterized protein-coding sequences-mostly from the intronic regions of known genes. Seventy-six of the 580 protein-coding genes were associated with cancer hallmark genes, which included key oncogenes, transcription factors, kinases, and cell surface receptors. Survival association analysis of the 76 novel peptide sequences revealed 10 of them to be significant, and we present a panel of six novel peptides, whose high expression was found to be strongly associated with poor survival of patients with human epidermal growth factor receptor 2-enriched subtype. Our analysis represents a landscape of novel peptides of different types that may be expressed in breast cancer tissues, whereas their presence in full-length functional proteins needs further investigations.
Keywords: CPTAC; alternative splicing; breast cancer; de novo transcript assembly; proteogenomics.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures




References
-
- Rusk N. From pseudogenes to proteins. Nat. Methods. 2011;8:448–449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

