HPV infection alters vaginal microbiome through down-regulating host mucosal innate peptides used by Lactobacilli as amino acid sources
- PMID: 35228537
- PMCID: PMC8885657
- DOI: 10.1038/s41467-022-28724-8
HPV infection alters vaginal microbiome through down-regulating host mucosal innate peptides used by Lactobacilli as amino acid sources
Abstract
Despite the high prevalence of both cervico-vaginal human papillomavirus (HPV) infection and bacterial vaginosis (BV) worldwide, their causal relationship remains unclear. While BV has been presumed to be a risk factor for HPV acquisition and related carcinogenesis for a long time, here, supported by both a large retrospective follow-up study (n = 6,085) and extensive in vivo data using the K14-HPV16 transgenic mouse model, we report a novel blueprint in which the opposite association also exists. Mechanistically, by interacting with several core members (NEMO, CK1 and β-TrCP) of both NF-κB and Wnt/β-catenin signaling pathways, we show that HPV E7 oncoprotein greatly inhibits host defense peptide expression. Physiologically secreted by the squamous mucosa lining the lower female genital tract, we demonstrate that some of these latter are fundamental factors governing host-microbial interactions. More specifically, several innate molecules down-regulated in case of HPV infection are hydrolyzed, internalized and used by the predominant Lactobacillus species as amino acid source sustaining their growth/survival. Collectively, this study reveals a new viral immune evasion strategy which, by its persistent/negative impact on lactic acid bacteria, ultimately causes the dysbiosis of vaginal microbiota.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


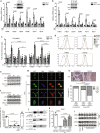

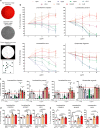
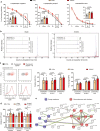


References
-
- de Sanjose S, et al. Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: a meta-analysis. Lancet Infect. Dis. 2007;7:453–459. - PubMed
-
- Schiffman M, et al. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Prim. 2016;2:16086. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

