Comparative systeomics to elucidate physiological differences between CHO and SP2/0 cell lines
- PMID: 35228567
- PMCID: PMC8885639
- DOI: 10.1038/s41598-022-06886-1
Comparative systeomics to elucidate physiological differences between CHO and SP2/0 cell lines
Abstract
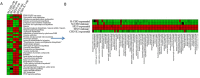
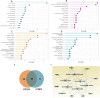
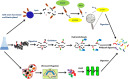
Omics-based tools were coupled with bioinformatics for a systeomics analysis of two biopharma cell types: Chinese hamster ovary (M-CHO and CHO-K1) and SP2/0. Exponential and stationary phase samples revealed more than 10,000 transcripts and 6000 proteins across these two manufacturing cell lines. A statistical comparison of transcriptomics and proteomics data identified downregulated genes involved in protein folding, protein synthesis and protein metabolism, including PPIA-cyclophilin A, HSPD1, and EIF3K, in M-CHO compared to SP2/0 while cell cycle and actin cytoskeleton genes were reduced in SP2/0. KEGG pathway comparisons revealed glycerolipids, glycosphingolipids, ABC transporters, calcium signaling, cell adhesion, and secretion pathways depleted in M-CHO while retinol metabolism was upregulated. KEGG and IPA also indicated apoptosis, RNA degradation, and proteosomes enriched in CHO stationary phase. Alternatively, gene ontology analysis revealed an underrepresentation in ion and potassium channel activities, membrane proteins, and secretory granules including Stxbpt2, Syt1, Syt9, and Cma1 proteins in M-CHO. Additional enrichment strategies involving ultracentrifugation, biotinylation, and hydrazide chemistry identified over 4000 potential CHO membrane and secretory proteins, yet many secretory and membrane proteins were still depleted. This systeomics pipeline has revealed bottlenecks and potential opportunities for cell line engineering in CHO and SP2/0 to improve their production capabilities.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

