PERK is a critical metabolic hub for immunosuppressive function in macrophages
- PMID: 35228694
- PMCID: PMC9112288
- DOI: 10.1038/s41590-022-01145-x
PERK is a critical metabolic hub for immunosuppressive function in macrophages
Abstract
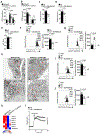
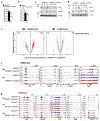
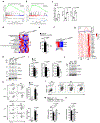
Chronic inflammation triggers compensatory immunosuppression to stop inflammation and minimize tissue damage. Studies have demonstrated that endoplasmic reticulum (ER) stress augments the suppressive phenotypes of immune cells; however, the molecular mechanisms underpinning this process and how it links to the metabolic reprogramming of immunosuppressive macrophages remain elusive. In the present study, we report that the helper T cell 2 cytokine interleukin-4 and the tumor microenvironment increase the activity of a protein kinase RNA-like ER kinase (PERK)-signaling cascade in macrophages and promote immunosuppressive M2 activation and proliferation. Loss of PERK signaling impeded mitochondrial respiration and lipid oxidation critical for M2 macrophages. PERK activation mediated the upregulation of phosphoserine aminotransferase 1 (PSAT1) and serine biosynthesis via the downstream transcription factor ATF-4. Increased serine biosynthesis resulted in enhanced mitochondrial function and α-ketoglutarate production required for JMJD3-dependent epigenetic modification. Inhibition of PERK suppressed macrophage immunosuppressive activity and could enhance the efficacy of immune checkpoint programmed cell death protein 1 inhibition in melanoma. Our findings delineate a previously undescribed connection between PERK signaling and PSAT1-mediated serine metabolism critical for promoting immunosuppressive function in M2 macrophages.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing financial interests
P.-C.H. is a member of scientific advisory for Elixiron Immunotherapeutics. The remaining authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

