Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes
- PMID: 35228745
- PMCID: PMC8938904
- DOI: 10.1038/s42255-022-00531-x
Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes
Abstract
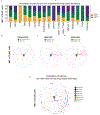
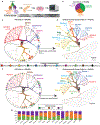
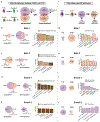
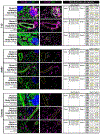
Type 1 diabetes (T1D) is an autoimmune disease in which immune cells destroy insulin-producing beta cells. The aetiology of this complex disease is dependent on the interplay of multiple heterogeneous cell types in the pancreatic environment. Here, we provide a single-cell atlas of pancreatic islets of 24 T1D, autoantibody-positive and nondiabetic organ donors across multiple quantitative modalities including ~80,000 cells using single-cell transcriptomics, ~7,000,000 cells using cytometry by time of flight and ~1,000,000 cells using in situ imaging mass cytometry. We develop an advanced integrative analytical strategy to assess pancreatic islets and identify canonical cell types. We show that a subset of exocrine ductal cells acquires a signature of tolerogenic dendritic cells in an apparent attempt at immune suppression in T1D donors. Our multimodal analyses delineate cell types and processes that may contribute to T1D immunopathogenesis and provide an integrative procedure for exploration and discovery of human pancreatic function.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

