Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress
- PMID: 35228749
- PMCID: PMC8885417
- DOI: 10.1038/s43018-022-00331-y
Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress
Abstract
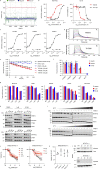
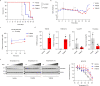
The folate metabolism enzyme MTHFD2 (methylenetetrahydrofolate dehydrogenase/cyclohydrolase) is consistently overexpressed in cancer but its roles are not fully characterized, and current candidate inhibitors have limited potency for clinical development. In the present study, we demonstrate a role for MTHFD2 in DNA replication and genomic stability in cancer cells, and perform a drug screen to identify potent and selective nanomolar MTHFD2 inhibitors; protein cocrystal structures demonstrated binding to the active site of MTHFD2 and target engagement. MTHFD2 inhibitors reduced replication fork speed and induced replication stress followed by S-phase arrest and apoptosis of acute myeloid leukemia cells in vitro and in vivo, with a therapeutic window spanning four orders of magnitude compared with nontumorigenic cells. Mechanistically, MTHFD2 inhibitors prevented thymidine production leading to misincorporation of uracil into DNA and replication stress. Overall, these results demonstrate a functional link between MTHFD2-dependent cancer metabolism and replication stress that can be exploited therapeutically with this new class of inhibitors.
© 2022. The Author(s).
Conflict of interest statement
C.B., S.B., M. Häggblad, T.H., M. Henriksson, E.H., C.P., L.S., M.S., P.S. and K.V. are named inventors on patent application PCT/EP2019/059919. MTHFD2 inhibitors are developed toward the clinic by the company One-Carbon Therapeutics AB, currently owned by The Helleday Foundation (THF), a not-for-profit charitable foundation. T.H. and U.W.B. are members of the board of THF. The remaining authors declare no competing interests.
Figures














Comment in
-
Beating cancer one carbon at a time.Nat Cancer. 2022 Feb;3(2):141-142. doi: 10.1038/s43018-022-00333-w. Nat Cancer. 2022. PMID: 35228750 No abstract available.
References
-
- Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–887. - PubMed
-
- Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. - PubMed
-
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. - PubMed
-
- Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. - PubMed
-
- Gorgoulis, V. G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature10.1038/nature03485 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

