Development of a hybrid alphavirus-SARS-CoV-2 pseudovirion for rapid quantification of neutralization antibodies and antiviral drugs
- PMID: 35229082
- PMCID: PMC8866097
- DOI: 10.1016/j.crmeth.2022.100181
Development of a hybrid alphavirus-SARS-CoV-2 pseudovirion for rapid quantification of neutralization antibodies and antiviral drugs
Abstract
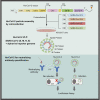
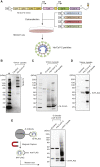
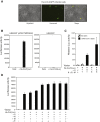
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein (S)-pseudotyped viruses are commonly used for quantifying antiviral drugs and neutralizing antibodies. Here, we describe the development of a hybrid alphavirus-SARS-CoV-2 (Ha-CoV-2) pseudovirion, which is a non-replicating SARS-CoV-2 virus-like particle composed of viral structural proteins (S, M, N, and E) and an RNA genome derived from a fast-expressing alphaviral vector. We validated Ha-CoV-2 for rapid quantification of neutralization antibodies, antiviral drugs, and viral variants. In addition, as a proof of concept, we used Ha-CoV-2 to quantify the neutralizing antibodies from an infected and vaccinated individual and found that the one-dose vaccination with Moderna mRNA-1273 greatly increased the anti-serum titer by approximately 6-fold. The post-vaccination serum can neutralize all nine variants tested. These results demonstrate that Ha-CoV-2 can be used as a robust platform for the rapid quantification of neutralizing antibodies against SARS-CoV-2 and its emerging variants.
Keywords: COVID-19; Ha-CoV-2; SARS-CoV-2; SARS-CoV-2 variants; alphavirus; antiviral drug; coronavirus; lentivirus; mRNA vaccine; neutralizing antibody; pseudovirus.
© 2022 The Author(s).
Conflict of interest statement
A patent application has been filed by George Mason University and is licensed for product development. Y.W. is a founder of Virongy Biosciences, Inc., and a member of its advisory board. T.C. is the chief scientific officer of Polaris Genomics, Inc.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

