USP25 inhibition ameliorates Alzheimer's pathology through the regulation of APP processing and Aβ generation
- PMID: 35229730
- PMCID: PMC8884900
- DOI: 10.1172/JCI152170
USP25 inhibition ameliorates Alzheimer's pathology through the regulation of APP processing and Aβ generation
Abstract
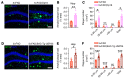
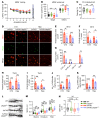
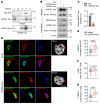
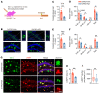
Down syndrome (DS), or trisomy 21, is one of the critical risk factors for early-onset Alzheimer's disease (AD), implicating key roles for chromosome 21-encoded genes in the pathogenesis of AD. We previously identified a role for the deubiquitinase USP25, encoded on chromosome 21, in regulating microglial homeostasis in the AD brain; however, whether USP25 affects amyloid pathology remains unknown. Here, by crossing 5×FAD AD and Dp16 DS mice, we observed that trisomy 21 exacerbated amyloid pathology in the 5×FAD brain. Moreover, bacterial artificial chromosome (BAC) transgene-mediated USP25 overexpression increased amyloid deposition in the 5×FAD mouse brain, whereas genetic deletion of Usp25 reduced amyloid deposition. Furthermore, our results demonstrate that USP25 promoted β cleavage of APP and Aβ generation by reducing the ubiquitination and lysosomal degradation of both APP and BACE1. Importantly, pharmacological inhibition of USP25 ameliorated amyloid pathology in the 5×FAD mouse brain. In summary, we identified the DS-related gene USP25 as a critical regulator of AD pathology, and our data suggest that USP25 serves as a potential pharmacological target for AD drug development.
Keywords: Alzheimer disease; Neuroscience.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

