XPG: a multitasking genome caretaker
- PMID: 35230528
- PMCID: PMC8888383
- DOI: 10.1007/s00018-022-04194-5
XPG: a multitasking genome caretaker
Abstract
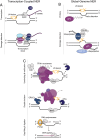
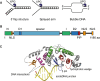
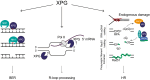
The XPG/ERCC5 endonuclease was originally identified as the causative gene for Xeroderma Pigmentosum complementation group G. Ever since its discovery, in depth biochemical, structural and cell biological studies have provided detailed mechanistic insight into its function in excising DNA damage in nucleotide excision repair, together with the ERCC1-XPF endonuclease. In recent years, it has become evident that XPG has additional important roles in genome maintenance that are independent of its function in NER, as XPG has been implicated in protecting replication forks by promoting homologous recombination as well as in resolving R-loops. Here, we provide an overview of the multitasking of XPG in genome maintenance, by describing in detail how its activity in NER is regulated and the evidence that points to important functions outside of NER. Furthermore, we present the various disease phenotypes associated with inherited XPG deficiency and discuss current ideas on how XPG deficiency leads to these different types of disease.
Keywords: DNA damage response; NER; Structure; XPG/ERCC5; Xeroderma pigmentosum–Cockayne syndrome.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.Proc Natl Acad Sci U S A. 2020 Jun 23;117(25):14127-14138. doi: 10.1073/pnas.1921311117. Epub 2020 Jun 10. Proc Natl Acad Sci U S A. 2020. PMID: 32522879 Free PMC article.
-
XPG in the Nucleotide Excision Repair and Beyond: a study on the different functional aspects of XPG and its associated diseases.Mol Biol Rep. 2022 Aug;49(8):7995-8006. doi: 10.1007/s11033-022-07324-1. Epub 2022 May 20. Mol Biol Rep. 2022. PMID: 35596054 Review.
-
A stable XPG protein is required for proper ribosome biogenesis: Insights on the phenotype of combinate Xeroderma Pigmentosum/Cockayne Syndrome patients.PLoS One. 2022 Jul 8;17(7):e0271246. doi: 10.1371/journal.pone.0271246. eCollection 2022. PLoS One. 2022. PMID: 35802638 Free PMC article.
-
Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene.Mol Cell Biol. 1999 Mar;19(3):2366-72. doi: 10.1128/MCB.19.3.2366. Mol Cell Biol. 1999. PMID: 10022922 Free PMC article.
-
Heterogeneity and overlaps in nucleotide excision repair disorders.Clin Genet. 2020 Jan;97(1):12-24. doi: 10.1111/cge.13545. Epub 2019 Apr 22. Clin Genet. 2020. PMID: 30919937 Review.
Cited by
-
Different SWI/SNF complexes coordinately promote R-loop- and RAD52-dependent transcription-coupled homologous recombination.Nucleic Acids Res. 2023 Sep 22;51(17):9055-9074. doi: 10.1093/nar/gkad609. Nucleic Acids Res. 2023. PMID: 37470997 Free PMC article.
-
Coordination of transcription-coupled repair and repair-independent release of lesion-stalled RNA polymerase II.Nat Commun. 2024 Aug 17;15(1):7089. doi: 10.1038/s41467-024-51463-x. Nat Commun. 2024. PMID: 39154022 Free PMC article.
-
ERCC5, HES6 and RORA are potential diagnostic markers of coronary artery disease.FEBS Open Bio. 2022 Oct;12(10):1814-1827. doi: 10.1002/2211-5463.13469. Epub 2022 Aug 7. FEBS Open Bio. 2022. PMID: 35934844 Free PMC article.
-
R-Loops in Genome Instability and Cancer.Cancers (Basel). 2023 Oct 14;15(20):4986. doi: 10.3390/cancers15204986. Cancers (Basel). 2023. PMID: 37894353 Free PMC article. Review.
-
Bridging the gap: R-loop mediated genomic instability and its implications in neurological diseases.Epigenomics. 2024 Mar 26;16(8):589-608. doi: 10.2217/epi-2023-0379. Online ahead of print. Epigenomics. 2024. PMID: 38530068 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

