Upregulation of antimicrobial peptide expression in slc26a3-/- mice with colonic dysbiosis and barrier defect
- PMID: 35230892
- PMCID: PMC8890434
- DOI: 10.1080/19490976.2022.2041943
Upregulation of antimicrobial peptide expression in slc26a3-/- mice with colonic dysbiosis and barrier defect
Abstract
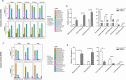
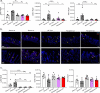
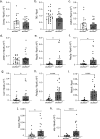
Genetic defects in SLC26A3 (DRA), an intestinal Cl-/HCO3- exchanger, result in congenital chloride diarrhea (CLD), marked by lifelong acidic diarrhea and a high risk of inflammatory bowel disease. Slc26a3-/- mice serve as a model to understand the pathophysiology of CLD and search for treatment options. This study investigates the microbiota changes in slc26a3-/- colon, the genotype-related causes for the observed microbiota alterations, its inflammatory potential, as well as the corresponding host responses. The luminal and the mucosa-adherent cecal and colonic microbiota of cohoused slc26a3-/- and wt littermates were analyzed by 16S rRNA gene sequencing. Fecal microbiota transfer from cohoused slc26a3-/- and wt littermates to germ-free wt mice was performed to analyze the stability and the inflammatory potential of the communities.The cecal and colonic luminal and mucosa-adherent microbiota of slc26a3-/- mice was abnormal from an early age, with a loss of diversity, of short-chain fatty acid producers, and an increase of pathobionts. The transfer of slc26a3-/- microbiota did not result in intestinal inflammation and the microbial diversity in the recipient mice normalized over time. A strong increase in the expression of Il22, Reg3β/γ, Relmβ, and other proteins with antimicrobial functions was observed in slc26a3-/- colon from juvenile age, while the mucosal and systemic inflammatory signature was surprisingly mild. The dysbiotic microbiota, low mucosal pH, and mucus barrier defect in slc26a3-/- colon are accompanied by a stark upregulation of the expression of a panel of antimicrobial proteins. This may explain the low inflammatory burden in the gut of these mice.
Keywords: Anion exchange; antimicrobial peptides; gut dysbiosis; inflammatory bowel disease; intestinal electrolyte transport.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
Escherichia coli Nissle Improves Short-Chain Fatty Acid Absorption and Barrier Function in a Mouse Model for Chronic Inflammatory Diarrhea.Inflamm Bowel Dis. 2025 Apr 10;31(4):1109-1120. doi: 10.1093/ibd/izae294. Inflamm Bowel Dis. 2025. PMID: 39708301 Free PMC article.
-
SLC26A3 (DRA, the Congenital Chloride Diarrhea Gene): A Novel Therapeutic Target for Diarrheal Diseases.Cell Mol Gastroenterol Hepatol. 2025;19(6):101452. doi: 10.1016/j.jcmgh.2024.101452. Epub 2024 Dec 28. Cell Mol Gastroenterol Hepatol. 2025. PMID: 39736385 Free PMC article. Review.
-
Slc26a3 deletion alters pH-microclimate, mucin biosynthesis, microbiome composition and increases the TNFα expression in murine colon.Acta Physiol (Oxf). 2020 Oct;230(2):e13498. doi: 10.1111/apha.13498. Epub 2020 Jun 20. Acta Physiol (Oxf). 2020. PMID: 32415725
-
Oral tributyrin treatment affects short-chain fatty acid transport, mucosal health, and microbiome in a mouse model of inflammatory diarrhea.J Nutr Biochem. 2025 Apr;138:109847. doi: 10.1016/j.jnutbio.2025.109847. Epub 2025 Jan 25. J Nutr Biochem. 2025. PMID: 39870330
-
Slc26a3 (DRA) in the Gut: Expression, Function, Regulation, Role in Infectious Diarrhea and Inflammatory Bowel Disease.Inflamm Bowel Dis. 2021 Mar 15;27(4):575-584. doi: 10.1093/ibd/izaa256. Inflamm Bowel Dis. 2021. PMID: 32989468 Review.
Cited by
-
SLC26 Anion Transporters.Handb Exp Pharmacol. 2024;283:319-360. doi: 10.1007/164_2023_698. Handb Exp Pharmacol. 2024. PMID: 37947907 Review.
-
Escherichia coli Nissle Improves Short-Chain Fatty Acid Absorption and Barrier Function in a Mouse Model for Chronic Inflammatory Diarrhea.Inflamm Bowel Dis. 2025 Apr 10;31(4):1109-1120. doi: 10.1093/ibd/izae294. Inflamm Bowel Dis. 2025. PMID: 39708301 Free PMC article.
-
SLC26A3 (DRA, the Congenital Chloride Diarrhea Gene): A Novel Therapeutic Target for Diarrheal Diseases.Cell Mol Gastroenterol Hepatol. 2025;19(6):101452. doi: 10.1016/j.jcmgh.2024.101452. Epub 2024 Dec 28. Cell Mol Gastroenterol Hepatol. 2025. PMID: 39736385 Free PMC article. Review.
-
Reduced surface pH and upregulated AE2 anion exchange in SLC26A3-deleted polarized intestinal epithelial cells.Am J Physiol Cell Physiol. 2024 Mar 1;326(3):C829-C842. doi: 10.1152/ajpcell.00590.2023. Epub 2024 Jan 15. Am J Physiol Cell Physiol. 2024. PMID: 38223928 Free PMC article.
-
The NHE3 Inhibitor Tenapanor Prevents Intestinal Obstructions in CFTR-Deleted Mice.Int J Mol Sci. 2022 Sep 1;23(17):9993. doi: 10.3390/ijms23179993. Int J Mol Sci. 2022. PMID: 36077390 Free PMC article.
References
-
- Camarillo GF, Goyon EI, Zuñiga RB, Salas LAS, Escárcega AEP, Yamamoto-Furusho JK. Gene expression profiling of mediators associated with the inflammatory pathways in the intestinal tissue from patients with ulcerative colitis. Mediators Inflamm. 2020;2020:9238970. doi: 10.1155/2020/9238970. - DOI - PMC - PubMed
-
- Yang H, Jiang W, Furth EE, Wen X, Katz JP, Sellon RK, Silberg DG, Antalis TM, Schweinfest CW, Wu GD. Intestinal inflammation reduces expression of DRA, a transporter responsible for congenital chloride diarrhea. Am J Physiol Liver Physiol. 1998;275(6):G1445–1453. doi: 10.1152/ajpgi.1998.275.6.G1445. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
