The FACT complex facilitates expression of lysosomal and antioxidant genes through binding to TFEB and TFE3
- PMID: 35230915
- PMCID: PMC9542721
- DOI: 10.1080/15548627.2022.2029671
The FACT complex facilitates expression of lysosomal and antioxidant genes through binding to TFEB and TFE3
Abstract
TFEB (transcription factor EB) and TFE3 (transcription factor binding to IGHM enhancer 3) orchestrate the cellular response to a variety of stressors, including nutrient deprivation, oxidative stress and pathogens. Here we describe a novel interaction of TFEB and TFE3 with the FAcilitates Chromatin Transcription (FACT) complex, a heterodimeric histone chaperone consisting of SSRP1 and SUPT16H that mediates nucleosome disassembly and assembly, thus facilitating transcription. Extracellular stimuli, such as nutrient deprivation or oxidative stress, induce nuclear translocation and activation of TFEB and TFE3, which then associate with the FACT complex to regulate stress-induced gene transcription. Depletion of FACT does not affect TFEB activation, stability, or binding to the promoter of target genes. In contrast, reduction of FACT levels by siRNA or treatment with the FACT inhibitor curaxin, severely impairs induction of numerous antioxidant and lysosomal genes, revealing a crucial role of FACT as a regulator of cellular homeostasis. Furthermore, upregulation of antioxidant genes induced by TFEB over-expression is significantly reduced by curaxin, consistent with a role of FACT as a TFEB transcriptional activator. Together, our data show that chromatin remodeling at the promoter of stress-responsive genes by FACT is important for efficient expression of TFEB and TFE3 targets, thus providing a link between environmental changes, chromatin modifications and transcriptional regulation.Abbreviations: ADNP2, ADNP homeobox 2; ATP6V0D1, ATPase H+ transporting V0 subunit d1; ATP6V1A, ATPase H+ transporting V1 subunit A; ATP6V1C1, ATPase H+ transporting V1 subunit C1; CSNK2/CK2, casein kinase 2; CLCN7, chloride voltage-gated channel 7; CTSD, cathepsin D; CTSZ, cathepsin Z; EBSS, earle's balanced salt solution; FACT complex, facilitates chromatin transcription complex; FOXO3, forkhead box O3; HEXA, hexosaminidase subunit alpha; HIF1A, hypoxia inducible factor 1 subunit alpha; HMOX1, heme oxygenase 1; LAMP1, lysosomal associated membrane protein 1; MAFF, MAF bZIP transcription factor F; MAFG, MAF bZIP transcription factor G; MCOLN1, mucolipin TRP cation channel 1; MTORC1, mechanistic target of rapamycin kinase complex 1; NaAsO2, sodium arsenite; POLR2, RNA polymerase II; PPARGC1A, PPARG coactivator 1 alpha; PYROXD1, pyridine nucleotide-disulfide oxidoreductase domain 1; RRAGC, Ras related GTP binding C; SEC13, SEC13 homolog, nuclear pore and COPII coat complex component; SLC38A9, solute carrier family 38 member 9; SSRP1, structure specific recognition protein 1; SUPT16H, SPT16 homolog, facilitates chromatin remodeling subunit; TFEB, transcription factor EB; TFE3, transcription factor binding to IGHM enhancer 3; TXNRD1, thioredoxin reductase 1; UVRAG, UV radiation resistance associated; WDR59, WD repeat domain 59.
Keywords: Autophagy; FACT; TFE3; TFEB; chaperone; histone; lysosomes.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
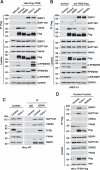

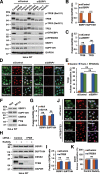
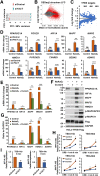
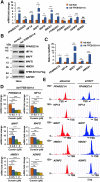
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
