Caspases and Their Substrates
- PMID: 35232877
- PMCID: PMC8886984
- DOI: 10.1101/cshperspect.a041012
Caspases and Their Substrates
Figures
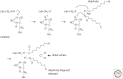

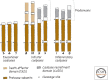

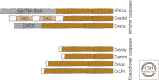






References
-
- Green DR. 2022c. Cell death and cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a041103 - DOI
FIGURE CREDITS
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
