Wildlife in Cameroon harbor diverse coronaviruses, including many closely related to human coronavirus 229E
- PMID: 35233291
- PMCID: PMC8867583
- DOI: 10.1093/ve/veab110
Wildlife in Cameroon harbor diverse coronaviruses, including many closely related to human coronavirus 229E
Abstract
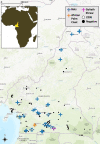
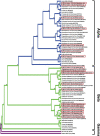
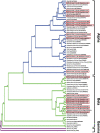
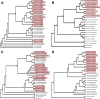
Zoonotic spillover of animal viruses into human populations is a continuous and increasing public health risk. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) highlights the global impact of emergence. Considering the history and diversity of coronaviruses (CoVs), especially in bats, SARS-CoV-2 will likely not be the last to spillover from animals into human populations. We sampled and tested wildlife in the Central African country Cameroon to determine which CoVs are circulating and how they relate to previously detected human and animal CoVs. We collected animal and ecological data at sampling locations and used family-level consensus PCR combined with amplicon sequencing for virus detection. Between 2003 and 2018, samples were collected from 6,580 animals of several different orders. CoV RNA was detected in 175 bats, a civet, and a shrew. The CoV RNAs detected in the bats represented 17 different genetic clusters, coinciding with alpha (n = 8) and beta (n = 9) CoVs. Sequences resembling human CoV-229E (HCoV-229E) were found in 40 Hipposideridae bats. Phylogenetic analyses place the human-derived HCoV-229E isolates closest to those from camels in terms of the S and N genes but closest to isolates from bats for the envelope, membrane, and RNA-dependent RNA polymerase genes. The CoV RNA positivity rate in bats varied significantly (P < 0.001) between the wet (8.2 per cent) and dry seasons (4.5 per cent). Most sampled species accordingly had a wet season high and dry season low, while for some the opposite was found. Eight of the suspected CoV species of which we detected RNA appear to be entirely novel CoV species, which suggests that CoV diversity in African wildlife is still rather poorly understood. The detection of multiple different variants of HCoV-229E-like viruses supports the bat reservoir hypothesis for this virus, with the phylogenetic results casting some doubt on camels as an intermediate host. The findings also support the previously proposed influence of ecological factors on CoV circulation, indicating a high level of underlying complexity to the viral ecology. These results indicate the importance of investing in surveillance activities among wild animals to detect all potential threats as well as sentinel surveillance among exposed humans to determine emerging threats.
Keywords: Coronavirus; HCoV-229E; bats; cameroon; seasonality; wildlife.
© The Author(s) 2021. Published by Oxford University Press.
Figures