Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain
- PMID: 35233548
- PMCID: PMC8784435
- DOI: 10.1016/j.xcrm.2022.100527
Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain
Abstract
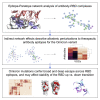
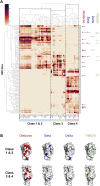
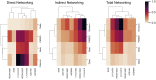
The Omicron variant features enhanced transmissibility and antibody escape. Here, we describe the Omicron receptor-binding domain (RBD) mutational landscape using amino acid interaction (AAI) networks, which are well suited for interrogating constellations of mutations that function in an epistatic manner. Using AAI, we map Omicron mutations directly and indirectly driving increased escape breadth and depth in class 1-4 antibody epitopes. Further, we present epitope networks for authorized therapeutic antibodies and assess perturbations to each antibody's epitope. Since our initial modeling following the identification of Omicron, these predictions have been realized by experimental findings of Omicron neutralization escape from therapeutic antibodies ADG20, AZD8895, and AZD1061. Importantly, the AAI predicted escape resulting from indirect epitope perturbations was not captured by previous sequence or point mutation analyses. Finally, for several Omicron RBD mutations, we find evidence for a plausible role in enhanced transmissibility via disruption of RBD-down conformational stability at the RBDdown-RBDdown interface.
Keywords: Omicron; RBD interface stability; SARS-CoV-2; antibody escape; antigenic drift; epistasis; epitope-paratope; mutation; network analysis; variant of concern.
© 2022 The Author(s).
Conflict of interest statement
R.S. is a board member of Tychan, Singapore, which focuses on infectious diseases.
Figures

Update of
-
Insights on the mutational landscape of the SARS-CoV-2 Omicron variant.bioRxiv [Preprint]. 2021 Dec 10:2021.12.06.471499. doi: 10.1101/2021.12.06.471499. bioRxiv. 2021. Update in: Cell Rep Med. 2022 Jan 24;3(2):100527. doi: 10.1016/j.xcrm.2022.100527. PMID: 34909771 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

